Validation of Biomarkers and Immunotherapy With Crohn's Disease Using WGCNA and Two-Sample Mendelian Randomization Study
- PMID: 40630894
- PMCID: PMC12237559
- DOI: 10.1155/grp/8194480
Validation of Biomarkers and Immunotherapy With Crohn's Disease Using WGCNA and Two-Sample Mendelian Randomization Study
Abstract
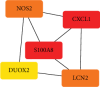
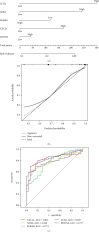
Objective: Crohn's disease (CD) is a chronic systemic inflammatory disease that mainly affects the intestine, accompanied by extraintestinal symptoms and immune problems. The progression of the disease may cause permanent damage to the structure and function of the intestine. Due to unclear early symptoms and lack of precise detection methods, early diagnosis of CD is difficult. Many patients were diagnosis at late stage, which may lead to delayed treatment and increased risk of complications. Identifying hub genes related to CD and using them to predict CD is of great significance. Methods: DEG and WGCNA were employed to identify key genes associated with CD and to detect modules significantly linked to the disease. GO and KEGG analyses were conducted to explore the functions of these identified genes. Additionally, MR method was utilized to assess the causal relationships between the most significant gene and CD. Results: WCGNA identified 3240 differentially expressed genes, with the magenta module being the most significant among the nine clustered modules. The enrichment of GO and KEGG pathways indicates that the hub genes in the magenta module are related to the positive regulation of heme binding, tetrapyrrole binding, carboxylic acid binding, organic acid binding, IL-17 signaling pathway, and amoebiasis pathway. The Top 5 hub genes are CXCL1, LCN2, NOS2, S100A8, and DUOX2. Mendelian randomization analysis found a significant correlation between CXCL1 and CD. Conclusions: The study screened five potential biomarker genes in CD patients using a bioinformatics approach and Mendelian randomization study. Our results provided insights into CXCL1, LCN2, NOS2, S100A8, and DUOX2 in CD and suggested that CXCL1 may potentially be the optimal biomarker that could be a relatively easy path to clinical translation.
Copyright © 2025 Cong Hu et al. Gastroenterology Research and Practice published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Interventions for the management of abdominal pain in Crohn's disease and inflammatory bowel disease.Cochrane Database Syst Rev. 2021 Nov 29;11(11):CD013531. doi: 10.1002/14651858.CD013531.pub2. Cochrane Database Syst Rev. 2021. PMID: 34844288 Free PMC article.
-
Intestinal mRNA expression profiles associated with mucosal healing in ustekinumab-treated Crohn's disease patients: bioinformatics analysis and prospective cohort validation.J Transl Med. 2024 Jun 26;22(1):595. doi: 10.1186/s12967-024-05427-w. J Transl Med. 2024. PMID: 38926732 Free PMC article.
-
A systematic review and economic evaluation of the use of tumour necrosis factor-alpha (TNF-α) inhibitors, adalimumab and infliximab, for Crohn's disease.Health Technol Assess. 2011 Feb;15(6):1-244. doi: 10.3310/hta15060. Health Technol Assess. 2011. PMID: 21291629 Free PMC article.
-
Uncovering common disease mechanisms and critical biomarkers in Crohn's disease with concurrent psoriasis and exploring potential therapeutic agents.PLoS One. 2025 Jun 20;20(6):e0324007. doi: 10.1371/journal.pone.0324007. eCollection 2025. PLoS One. 2025. PMID: 40540498 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

