This is a preprint.
Metabolic control of enteroendocrine cell fate through a redox state sensor CtBP
- PMID: 40631244
- PMCID: PMC12236740
- DOI: 10.1101/2025.06.30.662346
Metabolic control of enteroendocrine cell fate through a redox state sensor CtBP
Abstract
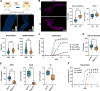
Enteroendocrine (EE) cells monitor the intestinal nutrient composition and consequently control organismal physiology through hormonal signaling. In addition to the immediate effects on hormone secretion, nutrients influence EE cell abundance by affecting the determination and maintenance of cell fate. EE cells are known to import and respond to dietary sugars, but how the sugar-induced changes in the intracellular metabolic state are sensed to control the immediate and long-term responses of EE cells, remains poorly understood. We report that the NADH binding transcriptional cofactor C-terminal binding protein (CtBP) acts at the interface between nutrient sensing and fate regulation of Drosophila larval EE cells, thus controlling organismal energy metabolism and survival on a high sugar diet. CtBP dimerization in EE cells is regulated through the redox balance of nicotinamide cofactors controlled by glycolysis and pentose phosphate pathway, allowing EE cells sense their internal metabolic state in response to sugar catabolism. CtBP interacts with the EE cell fate determining transcription factor Prospero through a conserved binding motif and binds to genomic targets controlling EE cell fate and size, such as components of Notch and insulin/mTOR pathways. Collectively, our findings uncover a modality where changes in intracellular redox state serve as an instructive signal to control EE cell function to globally control organismal homeostasis.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
