This is a preprint.
An Epigenomic Roadmap Primes Non-Growing Oocytes for Maturation and Early Embryogenesis
- PMID: 40631288
- PMCID: PMC12236492
- DOI: 10.1101/2025.07.06.663377
An Epigenomic Roadmap Primes Non-Growing Oocytes for Maturation and Early Embryogenesis
Abstract
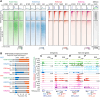
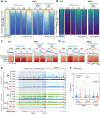
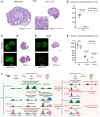
Female reproductive lifespan is defined by long-lived, non-growing oocytes (NGOs) that comprise the ovarian reserve. NGOs are assumed to acquire the epigenetic marks that will define the early embryo only after they exit the ovarian reserve and become activated for growth. Contrary to this dogma, we show that mouse NGOs possess abundant histone modifications that both underlie maintenance of the ovarian reserve and prime the epigenome of growing oocytes for early embryogenesis. As NGOs are established around birth, Polycomb Repressive Complex 1 (PRC1) mediates abundant H2AK119 ubiquitylation and reprograms the H3K27 acetylation landscape, which is essential to maintain the ovarian reserve. Importantly, the PRC1-driven epigenetic state of NGOs provides a blueprint for subsequent generation of a PRC2-catalyzed H3K27 trimethylation profile in growing oocytes that is characterized by broad domains and DNA methylation-independent imprints that are transmitted to the embryo. Thus, Polycomb complexes play pivotal roles in priming the NGO epigenome for oocyte maturation and early embryogenesis.
Keywords: Epigenetics; Epigenomics; Germline; Imprinting; Oogenesis; Ovarian reserve; Polycomb.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Ovarian biopsy has no role as a routine diagnostic test of ovarian reserve: a systematic review.Reprod Biomed Online. 2012 May;24(5):492-5. doi: 10.1016/j.rbmo.2012.01.020. Epub 2012 Jan 31. Reprod Biomed Online. 2012. PMID: 22414372
-
Individualised gonadotropin dose selection using markers of ovarian reserve for women undergoing in vitro fertilisation plus intracytoplasmic sperm injection (IVF/ICSI).Cochrane Database Syst Rev. 2018 Feb 1;2(2):CD012693. doi: 10.1002/14651858.CD012693.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2024 Jan 4;1:CD012693. doi: 10.1002/14651858.CD012693.pub3. PMID: 29388198 Free PMC article. Updated.
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
Human chorionic gonadotrophin priming for fertility treatment with in vitro maturation.Cochrane Database Syst Rev. 2016 Nov 16;11(11):CD008720. doi: 10.1002/14651858.CD008720.pub2. Cochrane Database Syst Rev. 2016. PMID: 27852101 Free PMC article.
-
Lower-order methylation states underlie the maintenance and re-establishment of Polycomb modifications in Drosophila embryogenesis.bioRxiv [Preprint]. 2025 Jul 29:2025.07.25.666882. doi: 10.1101/2025.07.25.666882. bioRxiv. 2025. PMID: 40766521 Free PMC article. Preprint.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
