Cellular Reprogramming by PHF7 Enhances Cardiac Function Following Myocardial Infarction
- PMID: 40631661
- PMCID: PMC12327843
- DOI: 10.1161/CIRCULATIONAHA.124.072733
Cellular Reprogramming by PHF7 Enhances Cardiac Function Following Myocardial Infarction
Abstract
Background: Direct reprogramming of fibroblasts to cardiomyocytes is a potentially curative strategy for ischemic heart disease. However, current reprogramming strategies require excessive factors due to epigenetic barriers of adult mouse and human fibroblasts. Recently, we identified the epigenetic factor PHF7 from a screen of gene-regulatory factors as the most potent activator of adult fibroblast-to-cardiomyocyte reprogramming in vitro.
Methods: Through in vitro assays coupled with genome-wide studies, we interrogated the ability of PHF7 to induce reprogramming events with minimal reprogramming factors. Using in vivo murine models of myocardial infarction and intramyocardial reprogramming factor delivery coupled with genetic fibroblast lineage tracing, we delivered retroviral PHF7 cocktails to the murine heart and interrogated reprogramming events as well as the acute and chronic functional impact of these cocktails. Deployment of 10X multiomics in vivo generated a combinatorial single-nucleus transcriptomic and epigenomic atlas of PHF7 reprogramming in the infarcted heart.
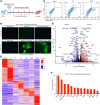
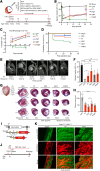
Results: Genome-wide in vitro transcriptomic analyses revealed that addition of PHF7 to Tbx5 or Mef2c and Tbx5 in fibroblasts induced global reprogramming through upregulation of unique cardiac transcriptomes. Further, PHF7 itself upregulated cardiac master regulators when overexpressed in dermal fibroblasts. Delivery of PHF7 cocktails to the infarcted murine heart induced in vivo reprogramming events and improved cardiac function and remodeling in both acute and chronic heart failure. When delivered as a single factor to the infarcted heart, PHF7 improved survival, function, and fibrosis up to 16 weeks after injury. Genetic lineage tracing analyses revealed that PHF7 induced bona fide fibroblast-to-cardiomyocyte reprogramming events in vivo. Comprehensive multiomics of PHF7 cocktails in the infarcted heart exposed the impact of PHF7 on chromatin structure, generating population-level shifts in nonmyocyte and cardiomyocyte cellular identity.
Conclusions: Here, we report the ability of a single epigenetic factor, PHF7, to induce reprogramming and improve cardiac function in the mouse heart following myocardial infarction. Together, these data support the premise that a single factor, when deployed into the infarcted mouse heart, can induce reprogramming events and recover function in the ischemic heart.
Keywords: cellular reprogramming; direct cell reprogramming techniques; epigenomics; heart failure; myocardial infarction.
Conflict of interest statement
Dr Olson is a cofounder and member of the scientific advisory board of Tenaya Therapeutics and holds equity in the company. Dr Olson is a consultant for Vertex Pharmaceuticals and Cardurion Pharmaceuticals. Drs Bann and Olson are inventors on US patent application 18/706 525 based on international patent application PCT/US2022/078854 entitled “Reprogramming of Adult Cardiac Fibroblasts Into Cardiomyocytes Using PHF7” in the name of the board of regents of the University of Texas System.
Figures


References
-
- Bozkurt B, Ahmad T, Alexander KM, Baker WL, Bosak K, Breathett K, Fonarow GC, Heidenreich P, Ho JE, Hsich E, et al. ; Writing Committee Members. Heart failure epidemiology and outcomes statistics: a report of the Heart Failure Society of America. J Card Fail. 2023;29:1412–1451. doi: 10.1016/j.cardfail.2023.07.006 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

