Crystal structures of Escherichia coli glucokinase and insights into phosphate binding
- PMID: 40632113
- PMCID: PMC12312565
- DOI: 10.1107/S2053230X25005515
Crystal structures of Escherichia coli glucokinase and insights into phosphate binding
Abstract
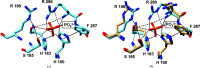
Here, we report the crystal structure of Escherichia coli glucokinase (GLK), which has phosphate bound in the cleft between the α and β domains adjacent to the active site. A ternary complex consisting of GLK, glucose and phosphate is also reported in this work. Diffraction data were collected at 2.63 Å resolution for the phospate-bound form (Rwork/Rfree = 0.191/0.230) and at 2.54 Å resolution for the ternary complex (Rwork/Rfree = 0.202/0.258), both at 297 K. A B-factor analysis of the phosphate-bound GLK structure revealed consistently lower values for phosphate-interacting basic residues in the α4, α5 and α9 helices, while significant root-mean-square deviation (r.m.s.d.) spikes indicated flexibility in regions preceding β1 and within the loop between the β5 and β6 sheets of the α domain. In the ternary complex, phosphate is bound adjacent to glucose, and the B factors for the α4, α5 and α9 helices were further reduced, while r.m.s.d. spikes were observed at the end of the β10 sheet and within the α6 helix of the β-domain. This structural characterization suggests that phosphate could influence the activity of GLK by altering glucose binding and modulating interactions with a loop-interacting regulatory protein.
Keywords: glucokinases; hexokinases; phosphate binding; sulfate binding.
open access.
Figures





Similar articles
-
Insights Into the Conformational Dynamics of the Cytoplasmic Domain of Metal-Sensing Sensor Histidine Kinase ZraS.Proteins. 2025 Sep;93(9):1465-1480. doi: 10.1002/prot.26819. Epub 2025 Mar 10. Proteins. 2025. PMID: 40062583 Free PMC article.
-
Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.J Bacteriol. 2004 Oct;186(20):6915-27. doi: 10.1128/JB.186.20.6915-6927.2004. J Bacteriol. 2004. PMID: 15466045 Free PMC article.
-
Isoleucine Binding and Regulation of Escherichia coli and Staphylococcus aureus Threonine Dehydratase (IlvA).Biochemistry. 2025 Jul 1;64(13):2793-2810. doi: 10.1021/acs.biochem.5c00168. Epub 2025 Jun 10. Biochemistry. 2025. PMID: 40494512 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

