Mitochondrial genome of eight Carangidae and phylogenetic analysis in the family
- PMID: 40632726
- PMCID: PMC12240318
- DOI: 10.1371/journal.pone.0326619
Mitochondrial genome of eight Carangidae and phylogenetic analysis in the family
Abstract
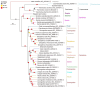
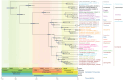
The Carangidae family is a prime focus for both deep-sea fishing and aquaculture. However, taxonomic controversies have limited Carangidae research. This study assembled the mitochondrial genomes of eight Carangidae species using second-generation sequencing and bioinformatics, then performed phylogenetic analyses. Mitochondrial genome sizes were: Megalaspis cordyla (16,565 bp; OR703829), Elagatis bipinnulata (16,543 bp; OR668919), Scomberoides tol (16,689 bp; OR668917), Selaroides leptolepis (16,560 bp; OR703831), Decapterus maruadsi (16,540 bp; OP459436), Alepes kleinii (16,570 bp; OR668918), Caranx sexfasciatus (16,595 bp; OR703830), and Carangoides orthogrammus (16,604 bp; OR668920). This study provides the first complete mitochondrial genome sequences of the species for Scomberoides tol, Carangoides orthogrammus, and Caranx sexfasciatus. The genomes contained two rRNA genes, 13 protein-coding genes, and 22-23 tRNAs, all with A + T bias. Phylogenetic analysis revealed a genetic distance of 0.002 between Uraspis secunda and U. helvola, suggesting that they are synonymous. The genetic distance between A. kleinii and A. djedaba was 0.082, reflecting their presence in the same genus. Intrageneric distance was greater than intergeneric distance between C. equula and C. orthogrammu, inconsistent with their taxonomic status. Finally, Seriolina and Caranginae were closely related, as were Trachinotinae and Chorineminae. In conclusion, our results provide breeding resources and an empirical basis for resolving Carangidae taxonomy.
Copyright: © 2025 Zhao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Nelson JS, Geande TC, Wilson MVH. Fishes of the World. Hoboken, New Jersey: John Wiley & Sons, Inc; 2016.
-
- Madduppa H, Martaulina R, Zairion Z, Renjani RM, Kawaroe M, Anggraini NP, et al. Genetic population subdivision of the blue swimming crab (Portunus pelagicus) across Indonesia inferred from mitochondrial DNA: Implication to sustainable fishery. PLoS One. 2021;16(2):e0240951. doi: 10.1371/journal.pone.0240951 - DOI - PMC - PubMed
-
- Ke X, Liu J, Gao F, Cao J, Liu Z, Lu M. Analysis of genetic diversity among six dojo loach (Misgurnus anguillicaudatus) populations in the Pearl River Basin based on microsatellite and mitochondrial DNA markers. Aquaculture Reports. 2022;27.
-
- Nielsen J, Rogers L, Brodeur R. Responses of ichthyoplankton assemblages to the recent marine heatwave and previous climate fluctuations in several Northeast Pacific marine ecosystems. Global Change Biology. 2020. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

