Clinical prediction of intravenous immunoglobulin-resistant Kawasaki disease based on interpretable Transformer model
- PMID: 40632807
- PMCID: PMC12240358
- DOI: 10.1371/journal.pone.0327564
Clinical prediction of intravenous immunoglobulin-resistant Kawasaki disease based on interpretable Transformer model
Abstract
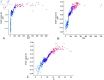
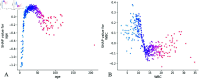
Intravenous immunoglobulin (IVIG) has been established as the first-line therapy for Kawasaki disease (KD). However, approximately 10%-20% of pediatric patients exhibit IVIG resistance. Current machine learning (ML) models demonstrate suboptimal predictive performance in KD treatment response prediction, primarily due to their limited ability to effectively process categorical variables and interpret tabular clinical data. This study aims to develop and interpretable transformer-based clinical prediction model for IVIG resistant KD and validate its clinical utility. This retrospective study analyzed clinical records of KD patients from the Affiliated Hospital of North Sichuan Medical College (Nanchong, China) between January 1, 2014 and December 31, 2024. A cohort of 1,578 pediatric KD cases was systematically divided into training and validation sets. Six machine learning algorithms - Random Forest (RF), AdaBoost, Light Gradient Boosting Machine (LightGBM), eXtreme Gradient Boosting (XGBoost), Categorical Boosting (CatBoost), and Tabular Prior-data Fitted Network version 2.0 (TabPFN-V2) - were implemented with five-fold cross-validation to optimize model hyperparameters. Model performance was rigorously evaluated using seven metrics: accuracy, precision, recall, F1-score, Matthews correlation coefficient (MCC), area under the receiver operating characteristic (ROC-AUC), and area under the precision-recall curve (PR-AUC). The top-performing model was subsequently subjected to interpretability analysis through Shapley Additive Explanations (SHAP) to elucidate feature contributions. The transformer-based TabPFN-V2 model demonstrated superior predictive performance in KD analysis, achieving an impressive validation set accuracy of 0.97. Comprehensive evaluation metrics confirmed its robust performance: precision 0.98, recall 0.97, F1-score 0.98, MCC 0.95, ROC-AUC 0.99, and PR-AUC 0.99. Global interpretability analysis through kernel SHAP methodology identified the ten most influential predictive features ranked by significance: Coronary artery lesions (CAL), Aspartate aminotransferase (AST), C-reactive protein (CRP), whether it was incomplete KD (KDtype), Neutrophil count (N), Platelet count (PLT), Albumin (ALB), age, White blood cell count (WBC) and Hemoglobin (Hb). Local interpretability analysis revealed distinct correlation patterns with IVIG resistance:AST, CRP, and N demonstrated significant positive correlations, where elevated values corresponded to increased IVIG resistance risk; PLT and ALB showed negative correlations, with higher levels associated with reduced resistance probability. Notably, age and WBC parameters demonstrated threshold effects, where optimal cutoff values enabled re-calibration of single-variable predictive scores. This threshold-dependent relationship suggests potential clinical utility in risk stratification protocols.The TabPFN-V2 model, leveraging an interpretable transformer architecture, demonstrates dual clinical utilities in KD management: (1) accurate prediction of IVIG resistance risk, and (2) data-driven support for personalized therapeutic decision-making. This framework enables probabilistic estimation of treatment resistance likelihood while providing transparent feature contribution analyses essential for developing patient-specific management protocols.
Copyright: © 2025 Chen, Yang. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Supervised Machine Learning Models for Predicting Sepsis-Associated Liver Injury in Patients With Sepsis: Development and Validation Study Based on a Multicenter Cohort Study.J Med Internet Res. 2025 May 26;27:e66733. doi: 10.2196/66733. J Med Internet Res. 2025. PMID: 40418571 Free PMC article.
-
Explainable deep learning algorithm for distinguishing IVIG-Resistant Kawasaki disease in Shandong peninsula, China.BMC Pediatr. 2025 Aug 28;25(1):658. doi: 10.1186/s12887-025-06082-w. BMC Pediatr. 2025. PMID: 40866882 Free PMC article.
-
Construction and validation of HBV-ACLF bacterial infection diagnosis model based on machine learning.BMC Infect Dis. 2025 Jul 1;25(1):847. doi: 10.1186/s12879-025-11199-5. BMC Infect Dis. 2025. PMID: 40596896 Free PMC article.
-
C-reactive protein to albumin ratio as a prognostic tool for predicting intravenous immunoglobulin resistance in children with kawasaki disease: a systematic review of cohort studies.Pediatr Rheumatol Online J. 2024 Apr 12;22(1):42. doi: 10.1186/s12969-024-00980-6. Pediatr Rheumatol Online J. 2024. PMID: 38610057 Free PMC article.
-
Learning-Based Models for Predicting IVIG Resistance and Coronary Artery Lesions in Kawasaki Disease: A Review of Technical Aspects and Study Features.Paediatr Drugs. 2025 Jul;27(4):465-479. doi: 10.1007/s40272-025-00693-7. Epub 2025 Apr 3. Paediatr Drugs. 2025. PMID: 40180759 Review.
References
-
- Shaanxi Provincial Diagnosis and Treatment Center of Kawasaki Disease, Clinical Research Center for Childhood Diseases of Shaanxi Province, Children’s Hospital of Shaanxi Provincial People’s Hospital, Editorial Board of Chinese Journal of Contemporary Pediatrics, Expert Committee of Advanced Training for Pediatrician, China Maternal andChildren’s Health Association, General Pediatrician Group, Society of Pediatrician, Chinese Doctor Association. Shaanxi provincial diagnosis and treatment center of Kawasaki Disease. Zhongguo Dang Dai Er Ke Za Zhi. 2021;23(9):867–76. doi: 10.7499/j.issn.1008-8830.2107110 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

