Spatiotemporal reconstruction of the North American A(H5N1) outbreak reveals successive lineage replacements by descendant reassortants
- PMID: 40632842
- PMCID: PMC12239939
- DOI: 10.1126/sciadv.adu4909
Spatiotemporal reconstruction of the North American A(H5N1) outbreak reveals successive lineage replacements by descendant reassortants
Abstract
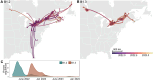
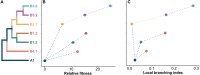
The November 2021 introduction of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b into North America triggered a devastating outbreak, affecting more than 180 million domestic birds and spreading to more than 80 wildlife species across Canada and the US. From this outbreak, we have sequenced 2955 complete A(H5N1) viral genomes from samples collected in Canada and, in conjunction with previously published data, performed multifaceted phylodynamic analyses. These analyses reveal extensive diversification of A(H5N1) viruses via reassortment with low-pathogenic avian influenza viruses. We find evidence of repeated ancestral strain replacement by direct descendants, indicative of compounding viral fitness increases. Spatiotemporal modeling identified critical geographic areas facilitating transcontinental spread and demonstrated genotype-specific host dynamics, offering essential data for ongoing control and prevention strategies.
Figures



References
-
- Olsen B., Munster V. J., Wallensten A., Waldenström J., Osterhaus A. D. M. E., Fouchier R. A. M., Global patterns of influenza A virus in wild birds. Science 312, 384–388 (2006). - PubMed
-
- S.-W. Yoon, R. J. Webby, R. G. Webster, “Evolution and ecology of influenza A viruses” in Influenza Pathogenesis and Control - Volume I (Springer International Publishing, 2014), pp. 359–375. - PubMed
-
- Venkatesh D., Poen M. J., Bestebroer T. M., Scheuer R. D., Vuong O., Chkhaidze M., Machablishvili A., Mamuchadze J., Ninua L., Fedorova N. B., Halpin R. A., Lin X., Ransier A., Stockwell T. B., Wentworth D. E., Kriti D., Dutta J., van Bakel H., Puranik A., Slomka M. J., Essen S., Brown I. H., Fouchier R. A. M., Lewis N. S., Avian influenza viruses in wild birds: Virus evolution in a multihost ecosystem. J. Virol. 92, e00433-18 (2018). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

