Synthetic efferocytic receptor microglia enhances anti-inflammatory clearance of amyloid-β for AD treatment in mice
- PMID: 40632863
- PMCID: PMC12239971
- DOI: 10.1126/sciadv.ads6613
Synthetic efferocytic receptor microglia enhances anti-inflammatory clearance of amyloid-β for AD treatment in mice
Abstract
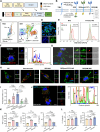
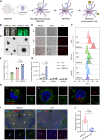
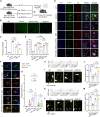
Monoclonal antibody immunotherapy targeting the clearance of amyloid-β (Aβ) has shown promise in Alzheimer's disease (AD). However, current antibody treatments trigger Fc receptors and induce proinflammatory responses, in turn exacerbating neuronal damage. Here, we report a synthetic efferocytic receptor (SER) integrating Aβ-targeting scFv, efferocytosis receptor backbone based on TIM4 and downstream signal for microglia (MG) reprogramming, which enabled selective elimination of Aβ without inducing an inflammatory response. Specifically, our in-house-customized MG-editing mRNA lipid nanoparticles (MERLINs) efficiently introduced SER mRNA into MG to generate Aβ-specific SER-MG in situ. SER-MG exhibited robust Aβ-specific phagocytosis and stimulated anti-inflammatory efferocytosis typical signaling in vitro. In a mouse model of AD, SER expression in the MG markedly increased the clearance of Aβ and dampened inflammation, resulting in improved behavioral outcomes along with substantially reduced synapse elimination. Our findings establish that AD-associated aberrant MG can be in situ reprogrammed with SER for Aβ clearance in an anti-inflammatory manner, with broad application in other inflammation-related diseases.
Figures



References
-
- Duyckaerts C., Delatour B., Potier M. C., Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 118, 5–36 (2009). - PubMed
-
- Kawas C., Gray S., Brookmeyer R., Fozard J., Zonderman A., Age-specific incidence rates of Alzheimer’s disease: The Baltimore longitudinal study of aging. Neurology 54, 2072–2077 (2000). - PubMed
-
- Masters C. L., Bateman R., Blennow K., Rowe C. C., Sperling R. A., Cummings J. L., Alzheimer’s disease. Nat. Rev. Dis. Primers 1, 15056 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

