GWAS meta-analysis identifies five susceptibility loci for endometrial cancer
- PMID: 40633141
- PMCID: PMC12275056
- DOI: 10.1016/j.ebiom.2025.105830
GWAS meta-analysis identifies five susceptibility loci for endometrial cancer
Abstract
Background: Endometrial cancer is the most common gynaecological cancer in high-income countries. In addition to environmental risk factors, genetic predisposition contributes towards endometrial cancer development but is still incompletely defined.
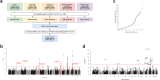
Methods: Building on genome-wide association studies (GWASs) by the Endometrial Cancer Association Consortium, we conducted a GWAS meta-analysis of 17,278 endometrial cancer cases and 289,180 controls, incorporating biobank samples from the UK, Finland, Estonia and Japan.
Findings: GWAS analysis identified five additional risk loci (3p25.2, 3q25.2, 6q22.31, 12q21.2, and 17q24.2). Corresponding gene-based analyses supported findings for three of the five loci, at NAV3 (12q21.2), PPARG (3p25.2), and BPTF (17q24.2), as well as two additional candidate risk regions at ATF7IP2 (16p13.2-p13.13) and RPP21 (6p22.1). Validation genotyping in further independent case-control series replicated the most significant locus at 12q21.2 and corroborated risk variants located intronic to NAV3, the gene for Neuron Navigator 3. Downregulation of NAV3 in endometrial cell lines accelerated cell division and wound healing capacity whereas NAV3 overexpression reduced cell survival and increased cell death, indicating that NAV3 acts as a tumour suppressor in endometrial cells.
Interpretation: Our large study extends the number of genome-wide significant risk loci identified for endometrial carcinoma by about one-third and proposes a role of NAV3 as a tumour suppressor in this common cancer.
Funding: This study was mainly supported by funding from the Wilhelm Sander Foundation, Germany, and the National Health and Medical Research Council (NHMRC) of Australia. A complete list of funding organisations is provided in the acknowledgements.
Keywords: Endometrial carcinoma; GWAS; Luciferase; NAV3; eQTL.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests D. Ramachandran received intramural funding from Hannover Medical School. M. Clark. has stock options in AbbVie and is employed by AbbVie on work not related to current manuscript. 1U01CA250476-01A1 to I. De Vivo and G. L. Mutter. NCI P30CA008748, U01CA250476 to M. Du. J. Emons receives honoraria for lectures from Pfizer, Eisai, AstraZeneca, Novartis, participates in the advisory board for these and MSD, receives travel money from AstraZeneca. Grant awards 3U01-CA199277-07S1 and 3U01-CA199277-08S1 from the NCI to J. V. Lacey for whole genome sequencing for participants in the California Teachers Study, which contributed data to this manuscript. P. A. Fasching receives grants from BioNTech, Cepheid, Pfizer; consulting fees, honoraria for lectures, and participates in an advisory board for Novartis, Pfizer, Roche, Daiichi-Sankyo, AstraZeneca, Lilly, Eisai, Merck Sharp & Dohme, Pierre Fabre, SeaGen, Agendia, Sanofi Aventis, Gilead, Mylan. D. L. Kolin holds stocks of Abbott laboratories, Alcon Inc., Becton Dickinson, Novartis, Pfizer, and UnitedHealth Group. G. L. Mutter received consulting fees from Bayer as personal consultant in pathology to perform diagnostic safety reads for clinical trials. A. V. Patel is on the NCI board of scientific counsellors. NIH funding to H. Risch. E. Tham received grants from Region Stockholm, the Swedish Childhood Cancer Fund (Barncancerfonden) and the Swedish Cancer Fund (Cancerfonden). E. Tham is a board member of Anna Dahlbäck’s memorial fund. Grants from CRUK and Genome Canada to D. Easton. NIH grant to P. Kraft. CRUK grants to P. Pharoah. D. Lambrechts receives annual funding from VIB. D. Glubb received payment from the University of Sharjah for grant reviewing. Grant from Wilhelm Sander Foundation to T. Dörk and P. Hillemanns. T. A. O’Mara received funding from the US Department of Defence and Worldwide Cancer Research. None of the sponsors had any role in the design, data generation or result interpretation in this study. All other authors declare no competing interests.
Figures




References
-
- Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. https://onlinelibrary.wiley.com/doi/10.3322/caac.21660 Available from: - DOI - PubMed
-
- Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21763 Available from: - DOI - PubMed
-
- Morice P., Leary A., Creutzberg C., Abu-Rustum N., Darai E. Endometrial cancer. Lancet. 2016;387(10023):1094–1108. https://linkinghub.elsevier.com/retrieve/pii/S0140673615001300 Available from: - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

