Feline infectious peritonitis epizootic caused by a recombinant coronavirus
- PMID: 40633571
- PMCID: PMC12408369
- DOI: 10.1038/s41586-025-09340-0
Feline infectious peritonitis epizootic caused by a recombinant coronavirus
Abstract
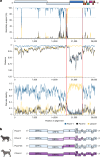
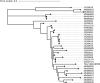
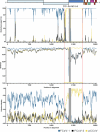
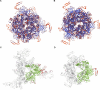
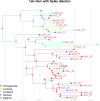
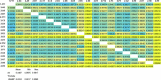
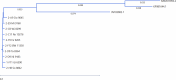
Cross-species transmission of coronaviruses (CoVs) poses a serious threat to both animal and human health1-3. While the large RNA genome of CoVs shows relatively low mutation rates, recombination within genera is frequently observed4-7. Companion animals are often overlooked in the transmission cycle of viral diseases; however, the close relationship of feline (FCoV) and canine CoV (CCoV) to human hCoV-229E5,8, as well as the susceptibility of these animals to SARS-CoV-29, highlight their importance in potential transmission cycles. While recombination between CCoV and FCoV of a large fragment spanning orf1b to M has been previously described5,10, here we report the emergence of a highly pathogenic FCoV-CCoV recombinant responsible for a rapidly spreading outbreak of feline infectious peritonitis (FIP) originating in Cyprus11. The minor recombinant region, spanning spike (S), shows 96.5% sequence identity to the pantropic canine coronavirus NA/09. Infection has rapidly spread, infecting cats of all ages. Development of FIP appears to be very frequent and sequence identities of samples from cats in different districts of the island are strongly supportive of direct transmission. A near-cat-specific deletion in the domain 0 of S is present in more than 90% of cats with FIP. It is unclear as yet whether this deletion is directly associated with disease development, and it may be linked to a biotype switch12. The domain 0 deletion and several amino acid changes in S, particularly the receptor-binding domain, indicate potential changes to receptor binding and cell tropism.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Laboklin is a veterinary laboratory offering diagnostic services, including bacteriological and molecular biological examinations. Vet Dia Gnosis offers veterinary pathology diagnostic services only. M.G. is employed by Laboklin. C.A. and S.L. are the co-founders of Vet Dia Gnosis. C.A. is an external collaborator of Vet Dia Gnosis and S.L. and A.Z. are employed by Vet Dia Gnosis.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

