Mosquito-borne alphaviruses in Zambia: Isolation and characterization of Eilat and Sindbis viruses
- PMID: 40633768
- PMCID: PMC12274731
- DOI: 10.1016/j.virusres.2025.199604
Mosquito-borne alphaviruses in Zambia: Isolation and characterization of Eilat and Sindbis viruses
Abstract
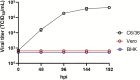
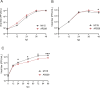
Alphaviruses in the family Togaviridae include zoonotic arthropod-borne viruses, including Sindbis virus (SINV), chikungunya virus, as well as insect-specific viruses such as Eilat virus (EILV). Previous investigations of alphaviruses in Zambia have identified a novel insect-specific alphavirus, Mwinilunga alphavirus in mosquitoes. Further ongoing surveillance resulted in the isolation of EILV and SINV for the first time in Zambia. Here, these alphaviruses were characterized in terms of growth kinetics in cells, and molecular phylogenetic relatedness to other alphaviruses. Zambian EILV (strain zmq19_M44) exhibited a close phylogenetic relationship with other insect-specific alphaviruses and shared a close nucleotide identity to those of EILV isolate (90.4 %) and Mwinilunga alphavirus (75.5 %). EILV zmq19_M44 attained a saturating titer in C6/36 cells at 6-8-days post infection but was unable to replicate in mammalian cells. Phylogenetic analysis revealed the Zambian SINV (strain zmq17_M115) belongs in Clade D of SINV Genotype 1 along with the Kenyan isolate BONI 584 from Central Africa. The growth of the SINV zmq17_M115 was comparable to that of the prototype SINV strain AR339 in mammalian cells but was statistically different in insect cells. Our findings will contribute to public health measures for the control of alphaviral diseases in Zambia.
Keywords: Alphavirus; Eilat virus; Mosquito; Sindbis virus; Zambia.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Eilat virus (EILV) causes superinfection exclusion against West Nile virus (WNV) in a strain-specific manner in Culex tarsalis mosquitoes.J Gen Virol. 2024 Aug;105(8):002017. doi: 10.1099/jgv.0.002017. J Gen Virol. 2024. PMID: 39189607
-
Pseudouridine synthases are proviral factors for Sindbis virus in insect and mammalian cells.mBio. 2025 Jul 9;16(7):e0132925. doi: 10.1128/mbio.01329-25. Epub 2025 May 27. mBio. 2025. PMID: 40422660 Free PMC article.
-
Insect-Specific Chimeric Viruses Potentiated Antiviral Responses and Inhibited Pathogenic Alphavirus Growth in Mosquito Cells.Microbiol Spectr. 2023 Feb 14;11(1):e0361322. doi: 10.1128/spectrum.03613-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511715 Free PMC article.
-
Significance of vertical transmission of arboviruses in mosquito-borne disease epidemiology.Parasit Vectors. 2025 Apr 9;18(1):137. doi: 10.1186/s13071-025-06761-8. Parasit Vectors. 2025. PMID: 40205559 Free PMC article.
-
Neuropathogenesis of Old World Alphaviruses: Considerations for the Development of Medical Countermeasures.Viruses. 2025 Feb 14;17(2):261. doi: 10.3390/v17020261. Viruses. 2025. PMID: 40007016 Free PMC article. Review.
References
-
- Adouchief S., Smura T., Sane J., Vapalahti O., Kurkela S. Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis. Rev. Med. Virol. 2016;26:221–241. - PubMed
-
- Babaniyi O.A., Mwaba P., Mulenga D., Monze M., Songolo P., Mazaba-Liwewe M.L., Mweene-Ndumba I., Masaninga F., Chizema E., Eshetu-Shibeshi M., Malama C., Rudatsikira E., Siziya S. Risk assessment for yellow fever in Western and north-Western provinces of zambia. J. Glob Infect Dis. 2015;7:11–17. - PMC - PubMed
-
- Bennouna A., Gil P., El Rhaffouli H., Exbrayat A., Loire E., Balenghien T., Chlyeh G., Gutierrez S., Fihri O.F. Identification of Eilat virus and prevalence of infection among Culex pipiens L. populations, Morocco, 2016. Virology. 2019;530:85–88. - PubMed
-
- Bergqvist J., Forsman O., Larsson P., Näslund J., Lilja T., Engdahl C., Lindström A., Gylfe Å., Ahlm C., Evander M., Bucht G. Detection and isolation of Sindbis virus from mosquitoes captured during an outbreak in Sweden, 2013. Vector Borne Zoonotic Dis. 2015;15:133–140. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

