M6A-METTL3-dependent nuclear PANC754/PSPC1/H3K4me1 repression complex regulate immune evasive LGALS7 signal to enhance immunotherapy against colorectal cancer
- PMID: 40634299
- PMCID: PMC12241656
- DOI: 10.1038/s41419-025-07820-9
M6A-METTL3-dependent nuclear PANC754/PSPC1/H3K4me1 repression complex regulate immune evasive LGALS7 signal to enhance immunotherapy against colorectal cancer
Abstract
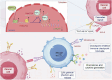
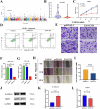
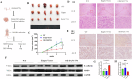
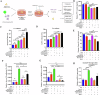
Non-coding RNAs (ncRNAs) have important regulatory functions similar to traditional oncogenes or tumor suppressor genes. Our previous research found a novel pan-cancer downexpressed ncRNA, PANC754. However, its function and underlying mechanism remain obscure in colorectal cancer (CRC). In this study, in vitro and in vivo experiments were performed to determine the function of PANC754. Loss and gain of function experiments, molecular docking experiments, and bioinformatic analysis were utilized to visualize its pathway. Co-culture system was leveraged to explore its effect on synergetic immune checkpoint blockage against CRC. Through a series of studies, we found that overexpressed PANC754 significantly inhibited cell viability, migration, and metastasis and induced notable apoptosis in CRC. The mechanical research found that PANC754 was the nuclear-located and its expression was regulated by m6A modification via METTL3 enzyme, which bound with its RBP PSPC1, then interacted with H3K4me1 to chromatin-accessible inhibit immune evasive molecule LGALS7 and led to suppress CRC progress. Furthermore, we confirmed that prominent upregulation of the immune checkpoint inhibitory (ICI) capability of anti-NKG2A, monalizumab when it was combined with PANC754 overexpression. Collectively, our study revealed that PANC754 is a tumor-suppressing ncRNA to form an ncRNA/RBP/histone repression complex with m6A-dependence, which can enhance the immune therapeutics effect of ICI, suggesting a promising therapeutic target. PANC754 is a novel pan-tumor suppressing non-coding RNA which is m6A-dependent and regulated by METTL3 modification enzyme. PANC754 is located at cellular nuclear and interacts with its RNA binding protein (RBP) PSPC1 and chromatin accessible histone H3K4me1, then can enhance immunotherapy capability of ICB anti-NKG2A against colorectal cancer through the immune evasive molecule LGALS7 signaling. Effect is that novel nuclear PANC754/PSPC1/H3K4me1 repression complex down-regulates the level "Don't eat me" signal LGALS7 to improve the immune efficiency of ICB and induce NK or CTL cell to release perforin and cytokine to kill tumor cells. (Created by Figdraw).
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The studies involving human participants were ethically reviewed and approved by the Institutional Review Board of Affiliated Hospital of Nantong University (No. 2018-K008). The patients/participants provided their written informed consent to participate in this study. The animal experiments were ethically inspected and approved by the Laboratory Animal Ethical Committee of Nantong University (No. S20210301-020).
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

