Clinical impact of pharmacogenetic risk variants in a large chinese cohort
- PMID: 40634300
- PMCID: PMC12241563
- DOI: 10.1038/s41467-025-61644-x
Clinical impact of pharmacogenetic risk variants in a large chinese cohort
Abstract
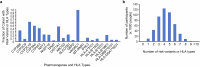
Incorporating pharmacogenetics into clinical practice promises to improve therapeutic outcomes by optimizing drug selection and dosage based on genetic factors affecting drug response. A key advantage of PGx-guided therapy is to decrease the likelihood of adverse events. To evaluate the clinical impact of PGx risk variants, we performed a retrospective study using genetic and clinical data from the largest Han Chinese cohort, comprising 486,956 individuals, assembled by the Taiwan Precision Medicine Initiative. We found that nearly all participants carried at least one genetic variant that could affect drug response, with many carrying multiple risk variants. Here we show the detailed analyses of four gene-drug pairs, azathioprine (NUDT15/TPMT), clopidogrel (CYP2C19), statins (ABCG2/CYP2C9/SLCO1B1), and NSAIDs (CYP2C9), for which sufficient data exists for statistical power. While the results validate previous findings that PGx risk variants are significantly associated with drug-related adverse events or ineffectiveness, the excess risk of adverse events or lack of efficacy is small compared to that found in those without the PGx risk variants, and most patients with PGx variants do not suffer from adverse events. Our results point to the complexity of implementing PGx in clinical practice and the need for integrative approaches to optimize precision medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Spear, B. B., Heath-Chiozzi, M. & Huff, J. Clinical application of pharmacogenetics. Trends Mol. Med7, 201–204 (2001). - PubMed
-
- Pirmohamed, M. Pharmacogenomics: current status and future perspectives. Nat. Rev. Genet24, 350–362 (2023). - PubMed
-
- Swen, J. J. et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet401, 347–356 (2023). - PubMed
-
- FDA. Table of Pharmacogenomic Biomarkers in Drug Labeling, https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenom.... Content current as of: 09/23/2024 (2024).

