A computational framework for defining and validating reproducible phenotyping algorithms of 313 diseases in the UK Biobank
- PMID: 40634319
- PMCID: PMC12241469
- DOI: 10.1038/s41598-025-05838-9
A computational framework for defining and validating reproducible phenotyping algorithms of 313 diseases in the UK Biobank
Abstract
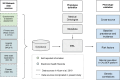
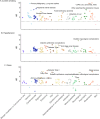
Accurate and reproducible phenotyping is essential for large-scale biomedical research. However, developing robust phenotype definitions in biobanks is challenging due to diverse data sources and varying medical ontologies. As a result, the current phenotyping landscape is fragmented. We developed a computational framework to harmonize electronic health record (EHR) data, participant questionnaires, and clinical registry information, defining 313 disease phenotypes among 502,356 UK Biobank (UKB) participants. Our method integrated four medical ontologies (Read v2, CTV3, ICD-10, OPCS-4) across seven data sources, including primary care, hospital admissions, cancer and death registries, and self-reported data on diseases, procedures, and medication. Phenotypes underwent multi-layered validation, assessing data source concordance, age-sex incidence and prevalence patterns, external comparison to a representative UK EHR dataset, modifiable risk factor associations, and genetic correlations with external genome-wide association studies (GWAS). Results indicated consistent disease distributions by age and sex, high correlation with non-selected general population data prevalence estimates, confirmed risk factor associations, and significant genetic correlations with external GWAS for nine of ten evaluated diseases. Our approach establishes comprehensive disease validation profiles, improving phenotype generalizability despite inherent UKB demographic biases. The modular, reproducible framework can be extended to additional diseases and populations, supporting federated analyses across diverse biobanks, and facilitating research in underrepresented populations.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: MGE, JCW, DCCC, ASC, TGR and JMD are employees and stakeholders of GlaxoSmithKline (GSK). JCW holds shares in GSK and was an employee of GSK until the end 2022. He has done extensive consultancy for pharma/biotech companies, with active agreements with Novo Nordisk, Relation Therapeutics Limited and Silence Therapeutics Plc. CT was employed at LifeArc, after manuscript preparation MB receives personal fees from GRAIL for membership of an Independent Data Monitoring Committee unrelated to this study. GF carried out work for this manuscript prior to starting as an employee at Novo Nordisk. The remaining authors have no conflict of interest.
Figures





References
-
- Nguyen, X.-M.T. et al. Data resource profile: Self-reported data in the million veteran program: Survey development and insights from the first 850 736 participants. Int. J. Epidemiol.52, 1–17. 10.1093/ije/dyac133 (2023). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

