Prediction and characterisation of the human B cell response to a heterologous two-dose Ebola vaccine
- PMID: 40634393
- PMCID: PMC12241346
- DOI: 10.1038/s41467-025-61571-x
Prediction and characterisation of the human B cell response to a heterologous two-dose Ebola vaccine
Abstract
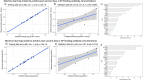
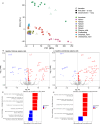
Ebola virus disease (EVD) outbreaks are increasing, posing significant threats to affected communities. Effective outbreak management depends on protecting frontline health workers, a key focus of EVD vaccination strategies. IgG specific to the viral glycoprotein serves as the correlate of protection for recent vaccine licensures. Using advanced cellular and transcriptomic analyses, we examined B cell responses to the Ad26.ZEBOV, MVA-BN-Filo EVD vaccine. Our findings reveal robust plasma cell and lasting B cell memory responses post-vaccination. Machine-learning models trained on blood gene expression predicted antibody response magnitude. Notably, we identified a unique B cell receptor CDRH3 sequence post-vaccination resembling known Orthoebolavirus zairense (EBOV) glycoprotein-binding antibodies. Single-cell analyses further detailed changes in plasma cell frequency, subclass usage, and CDRH3 properties. These results highlight the predictive power of early immune responses, captured through systems immunology, in shaping vaccine-induced B cell immunity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.J.P. is chair of the UK Department of Health and Social Care’s Joint Committee on Vaccination and Immunisation. A.J.P., S.B., E.A.C., K.A.S., R.M. and D.O.C. are contributors to intellectual property licensed by Oxford University Innovation to AstraZeneca. The remaining authors declare no competing interests.
Figures




References
-
- Henao-Restrepo, A. M. et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet386, 857–866 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

