Efficient secretion of a plastic degrading enzyme from the green algae Chlamydomonas reinhardtii
- PMID: 40634484
- PMCID: PMC12241367
- DOI: 10.1038/s41598-025-09100-0
Efficient secretion of a plastic degrading enzyme from the green algae Chlamydomonas reinhardtii
Abstract
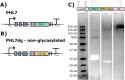
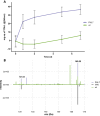
Plastic pollution has become a global crisis, with microplastics contaminating every environment on the planet, including our food, water, and even our bodies. In response, there is a growing interest in developing plastics that biodegrade naturally, thus avoiding the creation of persistent microplastics. As a mechanism to increase the rate of polyester plastic degradation, we examined the potential of using the green microalga Chlamydomonas reinhardtii for the expression and secretion of PHL7, an enzyme that breaks down post-consumer polyethylene terephthalate (PET) plastics. We engineered C. reinhardtii to secrete active PHL7 enzyme and selected strains showing robust expression, by using agar plates containing a polyester polyurethane (PU) dispersion as an efficient screening tool. This method demonstrated the enzyme's efficacy in degrading ester bond-containing plastics, such as PET and bio-based polyurethanes, and highlights the potential for microalgae to be implemented in environmental biotechnology. The effectiveness of algal-expressed PHL7 in degrading plastics was shown by incubating PET with the supernatant from engineered strains, resulting in substantial plastic degradation, confirmed by mass spectrometry analysis of terephthalic acid formation from PET. Our findings demonstrate the feasibility of polyester plastic recycling using microalgae to produce plastic-degrading enzymes. This eco-friendly approach can support global efforts toward eliminating plastic in our environment, and aligns with the pursuit of low-carbon materials, as these engineered algae can also produce plastic monomer precursors. Finally, this data demonstrates C. reinhardtii capabilities for recombinant enzyme production and secretion, offering a "green" alternative to traditional industrial enzyme production methods.
Keywords: Chlamydomonas reinhardtii; Climate-neutral economy; Environmental biotechnology; Microalgae; PETase; Secretion; Sustainable plastic recycling.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: SM was a founding member and holds an equity stake in Algenesis Materials Inc. MT and RS works at Algenesis Materials Inc. Algenesis Materials played no role in funding, study design, data collection and analysis, decision to publish, or manuscript preparation. This does not alter our adherence to policies on sharing data and materials. The remaining authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Sari, Y. W., Kartikasari, K., Widyarani, Setyaningsih, I. & Lestari, D. Chapter 13—Techno-economic assessment of microalgae for biofuel, chemical, and bioplastic. In Microalgae (ed. Galanakis, C. M.) 409–432 (Academic Press, 2021). 10.1016/B978-0-12-821218-9.00013-X.
-
- MacLeod, M., Arp, H. P. H., Tekman, M. B. & Jahnke, A. The global threat from plastic pollution. Science373, 61–65 (2021). - PubMed
-
- Cabernard, L., Pfister, S., Oberschelp, C. & Hellweg, S. Growing environmental footprint of plastics driven by coal combustion. Nat. Sustain.5, 139–148 (2021).
MeSH terms
Substances
Grants and funding
- DE-EE0009671/U.S. Department of Energy's Office of Energy Efficiency and Renewable Energy (EERE)
- DE-EE0009671/U.S. Department of Energy's Office of Energy Efficiency and Renewable Energy (EERE)
- DE-EE0009671/U.S. Department of Energy's Office of Energy Efficiency and Renewable Energy (EERE)
- DE-EE0009671/U.S. Department of Energy's Office of Energy Efficiency and Renewable Energy (EERE)
- DE-EE0009671/U.S. Department of Energy's Office of Energy Efficiency and Renewable Energy (EERE)
LinkOut - more resources
Full Text Sources
Research Materials

