A haplotype-resolved pangenome of the barley wild relative Hordeum bulbosum
- PMID: 40634612
- PMCID: PMC12422954
- DOI: 10.1038/s41586-025-09270-x
A haplotype-resolved pangenome of the barley wild relative Hordeum bulbosum
Abstract
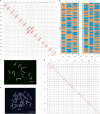
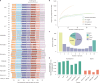
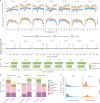
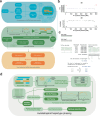
Wild plants can contribute valuable genes to their domesticated relatives1. Fertility barriers and a lack of genomic resources have hindered the effective use of crop-wild introgressions. Decades of research into barley's closest wild relative, Hordeum bulbosum, a grass native to the Mediterranean basin and Western Asia, have yet to manifest themselves in the release of a cultivar bearing alien genes2. Here we construct a pangenome of bulbous barley comprising 10 phased genome sequence assemblies amounting to 32 distinct haplotypes. Autotetraploid cytotypes, among which the donors of resistance-conferring introgressions are found, arose at least twice, and are connected among each other and to diploid forms through gene flow. The differential amplification of transposable elements after barley and H. bulbosum diverged from each other is responsible for genome size differences between them. We illustrate the translational value of our resource by mapping non-host resistance to a viral pathogen to a structurally diverse multigene cluster that has been implicated in diverse immune responses in wheat and barley.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



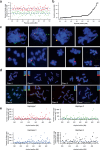



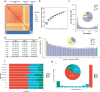

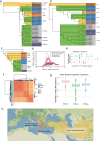


References
-
- Haas, M. & Mascher, M. Use of the secondary gene pool of barley in breeding improved varieties. Burleigh Dodds Chapters Online10.19103/AS.2019.0051.02 (2019).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

