Monitoring of antibodies against SARS-CoV-2 over 2 years and characterization of immune responses following Omicron infection in a South Indian community cohort
- PMID: 40634661
- PMCID: PMC12241409
- DOI: 10.1038/s41598-025-10447-7
Monitoring of antibodies against SARS-CoV-2 over 2 years and characterization of immune responses following Omicron infection in a South Indian community cohort
Abstract
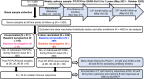
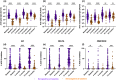
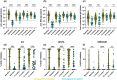
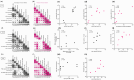
Multiple severe acute respiratory syndrome (SARS-CoV-2) exposures either due to infection or vaccination result in differential immune responses. Limited data exist on community-level SARS-CoV-2 variant exposures in India. We assessed antibody levels in 211 unvaccinated and 349 vaccinated individuals with and without prior exposure, followed for ~ 2 years (May 2021-October 2023) with baseline blood, weekly saliva for RT-PCR, and blood draws at ~ 6, 12, and 24 months. Humoral and cellular immune responses following primary (N = 8) and secondary (N = 11) Omicron infections in unvaccinated, previously exposed (N = 25), and unexposed vaccinated (N = 14) individuals were evaluated using Meso Scale Discovery and flow cytometry assays at Day 0, 30, and 90. Previously exposed individuals had higher anti-spike IgG levels at baseline (median 57,732 vs. 11,359, p < 0.001 in unvaccinated; 93,827 vs. 26,883, p < 0.001 in vaccinated) and 99% seropositivity at 2 years. Secondary Omicron infection elicited higher anti-spike IgG levels (17,320 vs. 3,930, p < 0.01) and stronger CD8 T cell responses (median CD8 AIM 0.026 vs. 0.001, p = 0.01 at Day90) than primary Omicron infection. Vaccinated individuals showed comparable CD4 and CD8 T cell responses irrespective of prior exposure, but previously exposed individuals had more durable antibodies at Day90 (p < 0.05 in previously exposed). Anti-spike IgG levels and %ACE-2 inhibition to Omicron were lower in both groups compared to other variants. High seropositivity was observed in both vaccinated and unvaccinated individuals after multiple exposures, though antibody levels were higher in individuals with prior exposure. These findings underscore preserved T cell responses to Omicron, although with lesser magnitude among unvaccinated.
Keywords: Antibody kinetics; Antigen imprinting; COVID-19 vaccination; Omicron variant.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The study has been approved by the Institutional Review Board of Christian Medical College, Vellore (IRB min no: 13585). We certify that the study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Consent to participate: Written informed consent was obtained from all participants.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

