Goblet cell breakdown: transcriptomics reveals Acinetobacter baumannii early and robust inflammatory response in differentiated human bronchial epithelial cells
- PMID: 40634947
- PMCID: PMC12239265
- DOI: 10.1186/s12929-025-01159-1
Goblet cell breakdown: transcriptomics reveals Acinetobacter baumannii early and robust inflammatory response in differentiated human bronchial epithelial cells
Abstract
Background: The airway epithelium represents the first line of defense of the lungs, functioning both as a physical barrier as well as an active immune modulator. However, in the last years, pneumonia caused by the opportunistic pathogen Acinetobacter baumannii have become difficult to treat due to the increase of the number of extensively drug resistant strains. In this study, we report for the first time the use of an ex vivo air-liquid interface (ALI) model of differentiated human bronchial epithelial cells to unravel the early response to A. baumannii infection.
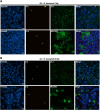
Methods: Epithelial integrity, tissue architecture, and goblet cell function were assessed through FITC-dextran permeability assays, hematoxylin and eosin staining, and indirect immunofluorescence. Transcriptomic profiling was performed to characterize host gene expression changes.
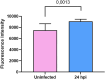
Results: Initial tissue damage began as early as at 4 h post-infection (hpi); at 24 hpi, goblet cell hypertrophy, reduced mucin secretion, and compromised epithelial integrity were highly evident. Transcriptomic data at 4 hpi revealed 668 differentially expressed genes (441 upregulated, 227 downregulated), mainly involved in a strong pro-inflammatory response and characterized by IL-8/CCL20-driven neutrophil recruitment and type 2 cytokine activation (IL-4, IL-13). Noteworthy, genes related to cytoskeletal organization, adhesion, and extracellular matrix remodeling were significantly altered, suggesting a bacterial mechanism to enhanced tissue dissemination. The PI3K-Akt survival pathway was inhibited, with downregulation of PIK3R1 and PIK3R2 genes, implying the induction of apoptosis/cell death and epithelial damage. Our findings are in agreement with previous in vivo studies, further strengthening the value of our ALI model in mimicking the early infection response of bronchial cells to A. baumannii infection.
Conclusion: Our data highlight the early molecular mechanisms underlying A. baumannii pathogenesis and open new avenues for future investigations for therapeutic interventions.
Keywords: Acinetobacter baumannii; 2D co-culture; Air liquid interface epithelium; Host–pathogen interaction; Infection model; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures





References
MeSH terms
Grants and funding
- Ricerca Corrente/Ministero della Salute
- RP12218162DFBF0A/Sapienza Università di Roma
- RP1221816196D9A2/Sapienza Università di Roma
- SEED PNR 2021 335/2021/Sapienza Università di Roma
- PRIN_2022 Prot. 2022WB59LB/Ministero dell'Università e della Ricerca
- PRIN_2022PNRR Prot. P2022C948R/Ministero dell'Università e della Ricerca
- NextGeneration EU-MUR M4C2.I.1.3 PNRR Extended Partnership initiative on Emerging Infectious Diseases (PE00000007/Ministero dell'Università e della Ricerca
- INF-ACT)/Ministero dell'Università e della Ricerca
- NextGeneration EU-MUR M4C2.I.1.3 PNRR Extended Partnership initiative on Emerging Infectious Diseases (PE00000007/Ministero dell'Università e della Ricerca
- INF-ACT)/Ministero dell'Università e della Ricerca
- MUR PRIN 2020 Prot. 2020KSY3KL/Ministero dell'Università e della Ricerca
- POR Lazio FSE 2014-202O/Regione Lazio
LinkOut - more resources
Full Text Sources
Miscellaneous

