Announcing the Biomedical Data Translator: Initial Public Release
- PMID: 40635371
- PMCID: PMC12241707
- DOI: 10.1111/cts.70284
Announcing the Biomedical Data Translator: Initial Public Release
Abstract
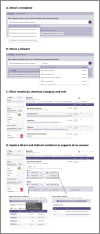
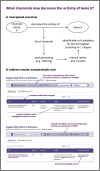
The growing availability of biomedical data offers vast potential to improve human health, but the complexity and lack of integration of these datasets often limit their utility. To address this, the Biomedical Data Translator Consortium has developed an open-source knowledge graph-based system-Translator-designed to integrate, harmonize, and make inferences over diverse biomedical data sources. We announce here Translator's initial public release and provide an overview of its architecture, standards, user interface, and core features. Translator employs a scalable, federated, knowledge graph framework for the integration of clinical, genomic, pharmacological, and other biomedical knowledge sources, enabling query retrieval, inference, and hypothesis generation. Translator's user interface is designed to support the exploration of knowledge relationships and the generation of insights, without requiring deep technical expertise and gradually revealing more detailed evidence, provenance, and confidence information, as needed by a given user. To demonstrate Translator's application and impact, we highlight features of the user interface in the context of three real-world use cases: suggesting potential therapeutics for patients with rare disease; explaining the mechanism of action of a pipeline drug; and screening and validating drug candidates in a model organism. We discuss strengths and limitations of reasoning within a largely federated system and the need for rich concept modeling and deep provenance tracking. Finally, we outline future directions for enhancing Translator's functionality and expanding its data sources. Translator represents a significant step forward in making complex biomedical knowledge more accessible and actionable, aiming to accelerate translational research and improve patient care.
© 2025 The Author(s). Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
S.E.B. and S.H. have received support from the NSF Convergence Accelerator Open Knowledge Networks to develop applications related to the SPOKE KG. All other authors declared no competing interests for this work.
Figures


References
-
- Wikipedia , “Knowledge Graph,” (2024), accessed November 21, 2024, https://en.wikipedia.org/w/index.php?title=Knowledge_graph&oldid=1255523142.

