Machine learning-based prediction of knee pain risk using lipid metabolism biomarkers: a prospective cohort study from CHARLS
- PMID: 40636147
- PMCID: PMC12239094
- DOI: 10.3389/fphys.2025.1607276
Machine learning-based prediction of knee pain risk using lipid metabolism biomarkers: a prospective cohort study from CHARLS
Abstract
Introduction: Knee pain significantly impairs health and quality of life among middle-aged and older adults. However, the predictive utility of lipid metabolism biomarkers for knee pain risk remains inadequately explored.
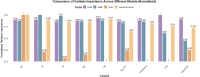
Methods: This study utilized data from the China Health and Retirement Longitudinal Study (CHARLS, 2011-2013) to investigate the association between lipid-related metabolic indicators and the risk of knee pain. Multiple lipid biomarkers and composite indices-including the lipid accumulation product (LAP), triglyceride-glucose (TyG) index, and TyG-BMI-were incorporated. Five machine learning models were developed and evaluated for predictive performance. Model interpretation was conducted using SHAP (SHapley Additive exPlanations) to identify the most influential predictors.
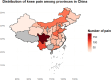
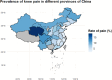
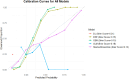
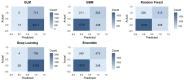
Results: A higher prevalence of knee pain was observed in high-altitude, cold regions such as Qinghai and Sichuan provinces. Composite metabolic indices (LAP, TyG, and TyG-BMI) exhibited stronger predictive power than traditional single lipid markers. Among the models, the Stacked Ensemble algorithm achieved the best performance, with an AUC of 0.85 and a Brier score of 0.13. SHAP analysis highlighted LAP and TyG-related indices as the top contributors to prediction outcomes.
Discussion: These findings emphasize the importance of lipid metabolism indicators in the early identification of knee pain risk. The integration of interpretable machine learning approaches and composite metabolic indices offers a promising strategy for personalized prevention in aging populations.
Keywords: CHARLS; knee pain; lipid accumulation product; machine learning; metabolic biomarkers.
Copyright © 2025 Guo, Li, Peng, Liu, He and Zhai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The value of triglyceride-glucose index-related indices in evaluating migraine: perspectives from multi-centre cross-sectional studies and machine learning models.Lipids Health Dis. 2025 Jul 3;24(1):230. doi: 10.1186/s12944-025-02648-w. Lipids Health Dis. 2025. PMID: 40611268 Free PMC article.
-
Multimodal machine learning-based marker enables the link between obesity-related indices and future stroke: a prospective cohort study.EClinicalMedicine. 2025 Jul 1;85:103331. doi: 10.1016/j.eclinm.2025.103331. eCollection 2025 Jul. EClinicalMedicine. 2025. PMID: 40678699 Free PMC article.
-
The association of obesity and lipid-related indicators with all-cause and cardiovascular mortality risks in patients with diabetes or prediabetes: a cross-sectional study based on machine learning algorithms.Front Endocrinol (Lausanne). 2025 Jun 2;16:1492082. doi: 10.3389/fendo.2025.1492082. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40529828 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Exercise interventions and patient beliefs for people with hip, knee or hip and knee osteoarthritis: a mixed methods review.Cochrane Database Syst Rev. 2018 Apr 17;4(4):CD010842. doi: 10.1002/14651858.CD010842.pub2. Cochrane Database Syst Rev. 2018. PMID: 29664187 Free PMC article.
References
-
- Chawla N. V., Bowyer K. W., Hall L. O., Kegelmeyer W. P. (2002). SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357. 10.1613/jair.953 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

