ApoE3 Christchurch and tau interaction as a protective mechanism against Alzheimer's disease
- PMID: 40637118
- PMCID: PMC12242688
- DOI: 10.1002/alz.70396
ApoE3 Christchurch and tau interaction as a protective mechanism against Alzheimer's disease
Abstract
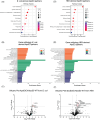
Introduction: We described a protected case with familial Alzheimer's disease, homozygous for apolipoprotein E3 (APOE3) Christchurch variant (ApoE3Ch), exhibiting low tau protein levels despite genetic predisposition to the disease due to presenilin (PSEN)1-E280A. We reported the loss of interaction between ApoE3Ch and heparan sulfate proteoglycans (HSPGs) as a critical protective pathway. Here, we characterized differential interacting partners for both wild-type and Christchurch variants to identify additional protective mechanisms of ApoE3Ch.
Methods: We performed pull-down of mouse brain lysates using His-tag-ApoE3 recombinant proteins and determined interacting partners of ApoE3 via mass-spectrometry. We then performed in vitro and in vivo assays to validate the top interactors.
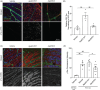
Results: We found enhanced binding of ApoE3Ch to tau and Dickkopf-1 (Dkk1, a WNT/β-catenin antagonist) that resulted in reduced tau aggregation in vitro. We demonstrated that ApoE3Ch interacts directly with Dkk1 and tau, reducing tau pathology. These findings supported the hypothesis of novel protective effects of direct ApoE3Ch interactions.
Highlights: Apolipoprotein E3 (ApoE3) Christchurch variant (ApoE3Ch) exhibits different protein interaction profiles compared to wild-type ApoE3, as revealed by proteomic analyses and pull-down experiments. The ApoE3Ch variant alters the protein's interaction with tau, thus affecting its aggregation in a tau biosensor cell assay and the retina of microtubule-associated protein tau (MAPT*P301S) transgenic mice. Gene ontology and pathway analyses indicate that ApoE3Ch interactors are associated with brain-related disorders and specific upstream regulators, including MAPT, a gene encoding for tau. Protein-protein interaction studies showed increased binding of ApoE3Ch to Dickkopf1 (Dkk1), a Wnt/β-catenin pathway antagonist, as compared to ApoE3WT, thus indicating that multiple protective mechanisms are regulated by the ApoE3Ch variant Our study uncovers a novel protective effect of the ApoE3Ch variant against tau pathology, thus proposing new insights into Alzheimer's disease mechanisms and potential therapeutic targets.
Keywords: Alzheimer's disease; ApoE3 Christchurch; Dkk1; apolipoprotein E; oligomerization; tau.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Dr. J. Arboleda‐Velasquez is listed as co‐inventor on a patent to leverage therapeutics based on the ApoE3 Christchurch findings filed by Mass General Brigham. Drs. Arboleda‐Velasquez and Kim are co‐founders of Epoch Biotech, a company focused on developing therapeutics inspired by the Christchurch variant. All other authors have no conflicts to disclose. Author disclosures are available in the Supporting Information.
Figures



References
-
- Alzheimer's Association . 2019 Alzheimer's disease facts and figures. Alzheimer's Association; 2019.
-
- Acosta‐Baena N, Sepulveda‐Falla D, Lopera‐Gomez CM, et al. Pre‐dementia clinical stages in presenilin 1 E280A familial early‐onset Alzheimer's disease: a retrospective cohort study. Lancet Neurol. 2011;10(3):213‐220. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

