Key RNA-binding domains in the La protein establish tRNA modification levels in Trypanosoma brucei
- PMID: 40637228
- PMCID: PMC12242769
- DOI: 10.1093/nar/gkaf594
Key RNA-binding domains in the La protein establish tRNA modification levels in Trypanosoma brucei
Abstract
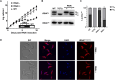
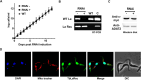
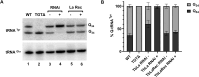
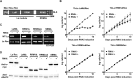
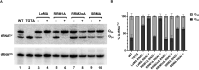
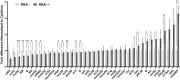
The RNA-binding protein La is found in most eukaryotes, and despite being essential in many organisms, its function is not completely clear. Trypanosoma brucei, the causative agent of human African trypanosomiasis, encodes a 'classical' La protein (TbLa) composed of a La-motif, two RNA recognition motifs (RRM1 and RRM2α), a C-terminal short basic motif (SBM), and a nuclear localization signal (NLS). In T. brucei, like in most eukaryotes, position 34 of tRNATyr, -Asp, -Asn and -His is modified with queuosine (Q34). The steady-state levels of queuosine-modified tRNA in the insect form (procyclic) of T. brucei can fluctuate dynamically depending on growth conditions, but the mechanism(s) controlling Q34 levels are not well understood. A well-established function of La is in precursor-tRNA 3'-end metabolism, but in this work, we demonstrate that La also controls Q34-tRNA levels. Individual domain deletions showed that while deletion of La motif or RRM1 causes dysregulation of Q34-tRNA levels, no other domain plays a similar role. We also show that La is important for the normal balance of several additional tRNA modifications. These findings are discussed in the context of substrate competition between La and modification enzymes, also highlighting subcellular localization as a key determinant of tRNA function.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

