Dissecting the epigenetic regulation of the fetal hemoglobin genes to unravel a novel therapeutic approach for β-hemoglobinopathies
- PMID: 40637230
- PMCID: PMC12242770
- DOI: 10.1093/nar/gkaf637
Dissecting the epigenetic regulation of the fetal hemoglobin genes to unravel a novel therapeutic approach for β-hemoglobinopathies
Abstract
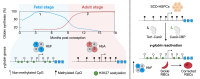
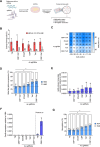
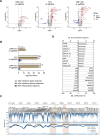
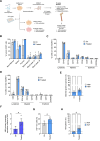
Beta-hemoglobinopathies are severe genetic diseases caused by mutations affecting the production of the adult β-globin chain. The clinical severity is mitigated by the co-inheritance of mutations that reactivate the production of the fetal β-like γ-globin in adults. However, the epigenetic mechanisms underlying the adult-to-fetal hemoglobin (HbA-to-HbF) switching are still not fully understood. Here, we used epigenome editing technologies to dissect the molecular mechanisms underlying γ- and β-globin gene regulation and to develop novel potential therapeutics for β-hemoglobinopathies. Targeted removal of DNA methylation by dCas9-Tet1 (alone or together with the deposition of histone acetylation by CBP-dCas9) at the fetal promoters led to efficient and durable γ-globin reactivation, demonstrating that DNA methylation is a driver for HbF repression. This strategy, characterized by high specificity and a good safety profile, led to a substantial correction of the pathological phenotype in erythroid cells from patients with sickle cell disease.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
S.A., L.F., and A.M. are named as inventors on a patent describing epigenome editing approaches for hemoglobinopathies (EP24315519.9: epigenetic reactivation of gamma-globin expression as a novel curative option for β-hemoglobinopathies). The remaining authors declare no competing interests.
Figures


References
-
- Kato GJ, Piel FB, Reid CD et al. Sickle cell disease. Nat Rev Dis Primer. 2018; 4:1–22. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

