Telomeric repeat-containing RNA increases in aged human cells
- PMID: 40637232
- PMCID: PMC12242772
- DOI: 10.1093/nar/gkaf597
Telomeric repeat-containing RNA increases in aged human cells
Abstract
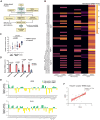
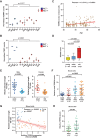
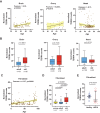
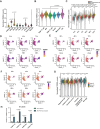
Telomeric repeat-containing RNA (TERRA), transcribed from subtelomeric regions toward telomeric ends, poses challenges in deciphering its complete sequences. Utilizing TERRA-capture RNA-seq and Oxford Nanopore direct RNA sequencing to acquire full-length TERRA, we annotate TERRA transcription regions in the human T2T-CHM13 reference genome. TERRA transcripts encompass hundreds to over a thousand nucleotides of telomeric repeats, predominantly originating from 61-29-37 bp repeat promoters enriched with H3K4me3, RNA Pol II, CTCF, and R-loops. We develop a bioinformatics tool, TERRA-QUANT, for quantifying TERRA using RNA-seq datasets and find that TERRA increases with age in blood, brain, and fibroblasts. TERRA upregulation in aged leukocytes is confirmed by reverse transcription quantitative polymerase chain reaction. Single-cell RNA-seq analysis demonstrates TERRA expression across various cell types, with upregulation observed in neurons during human embryonic stem cell differentiation. Additionally, TERRA levels are elevated in brain cells in the early stage of Alzheimer's disease. Our study provides evidence linking TERRA to human aging and diseases.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures




References
MeSH terms
Substances
Grants and funding
- NSTC 112-2628-B-002-008/National Science and Technology Council
- NSTC 112-2320-B-002-058/National Science and Technology Council
- NSTC 113-2320-B-002-009/National Science and Technology Council
- NSTC 113-2628-B-002-010-MY3/National Science and Technology Council
- NTU-111L7880/National Taiwan University
- NTU-AS-112L104312/National Taiwan University
- NTU-CDP-112L7721/National Taiwan University
- NTU-CDP-113L7705/National Taiwan University
- H-P.C.C./National Taiwan University
- NHRI-EX111-11107SI/National Health Research Institutes
- NHRI-EX112-11107SI/National Health Research Institutes
- NHRI-EX113-11107SI/National Health Research Institutes
- NHRI-EX114-11411SI/National Health Research Institutes
LinkOut - more resources
Full Text Sources
Medical

