Nucleolar FRG2 lncRNAs inhibit rRNA transcription and cytoplasmic translation, linking FSHD to dysregulation of muscle-specific protein synthesis
- PMID: 40637237
- PMCID: PMC12242771
- DOI: 10.1093/nar/gkaf643
Nucleolar FRG2 lncRNAs inhibit rRNA transcription and cytoplasmic translation, linking FSHD to dysregulation of muscle-specific protein synthesis
Abstract
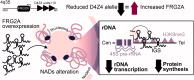
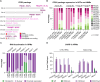
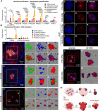
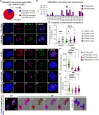
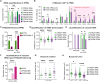
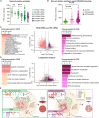
Facioscapulohumeral muscular dystrophy (FSHD) is a hereditary myopathy linked to deletions of the tandemly arrayed D4Z4 macrosatellite at human chromosome 4q35. These deletions cause local chromatin changes and anomalous expression of nearby transcripts FRG2A, DBET, and D4Z4. We discovered that FRG2A is part of a family of long noncoding RNAs (lncRNAs) expressed in skeletal muscle cells, with levels varying among patients. FRG2A localizes in the nucleolus and associates with repetitive DNA at ribosomal DNA (rDNA) loci and centromeres. Elevated FRG2A expression in FSHD cells alters the three-dimensional architecture of heterochromatin at the nucleolar periphery and reduces rDNA transcription and translation rates, resulting in decreased synthesis of skeletal muscle proteins. We also show that myoblasts from FSHD patients display reduced synthesis of skeletal muscle proteins during differentiation. Our results support a disease model in which nucleolar accumulation of D4Z4-driven lncRNA impairs protein synthesis and contributes to muscle wasting.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Winokur ST, Bengtsson U, Feddersen J et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994; 2:225–34. 10.1007/BF01553323. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- European Union -NextGenerationEU
- Italian Ministry of University
- M4C2-I1.3/Research under PNRR
- PE_00000019/Research under PNRR
- E93C22001860006/Research under PNRR
- University of Modena and Reggio Emilia
- RO1NS0475840/NH/NIH HHS/United States
- U01 CA260669/NH/NIH HHS/United States
- R35 GM15220/NH/NIH HHS/United States
- 1/NH/NIH HHS/United States
- FSHD Global Research Foundation
- GJC23060/Cariplo-Telethon Alliance
- EJ-2022-23683266/INNOVA
- IR0000010/INNOVA
- Departments of Excellence
- GJC23060/Cariplo-Telethon Alliance
LinkOut - more resources
Full Text Sources

