AREG and EREG Are Predictive Biomarkers of Response to EGFR Inhibition in Gastroesophageal Cancer
- PMID: 40637454
- PMCID: PMC12355176
- DOI: 10.1158/0008-5472.CAN-25-0073
AREG and EREG Are Predictive Biomarkers of Response to EGFR Inhibition in Gastroesophageal Cancer
Abstract
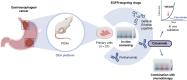
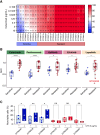
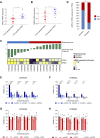
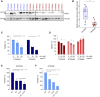
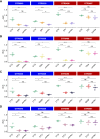
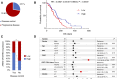
EGFR is a potential therapeutic target in gastroesophageal cancer. However, negative results from several phase II/III clinical trials have hindered the approval of EGFR inhibitors for treating gastroesophageal adenocarcinoma. Preclinical and clinical results have shown that EGFR targeting is effective in patients with gastroesophageal adenocarcinoma harboring EGFR amplification. Retrospective analyses also suggest that a subset of patients with gastroesophageal adenocarcinoma lacking EGFR amplification may benefit from the treatment, thus underscoring the need to identify reliable predictive biomarkers of response. Through the screening of 27 gastroesophageal adenocarcinoma primary cancer cell lines and 10 patient-derived xenograft models, we identified a subset of gastroesophageal adenocarcinoma lacking EGFR quantitative alterations but sensitive to EGFR targeting. Molecular characterization of the sensitive models revealed overexpression of the EGFR ligand amphiregulin (AREG) or epiregulin (EREG). Post hoc analysis of patients on the Cancer Esophagus Gefitinib trial treated with the EGFR inhibitor gefitinib demonstrated a significant correlation between overall survival and AREG/EREG expression level. No predictive power of EGFR ligand expression was observed in the presence of KRAS mutations. In conclusion, this study proposes the existence of a subgroup of patients with gastroesophageal adenocarcinoma with susceptibility to EGFR inhibition driven by overexpression of the EGFR ligands AREG and EREG.
Significance: Elevated levels of AREG or EREG in gastroesophageal cancer confers sensitivity to EGFR inhibition, providing a low-toxicity treatment option for the subpopulation of patients overexpressing the EGFR ligands.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
F. Pietrantonio reports receiving research funding (to institution) from Eli Lilly and Company, Bristol Myers Squibb, Incyte, AstraZeneca, Amgen, Agenus, and Rottapharm Biotech, personal honoraria as an invited speaker from BeiGene, Daiichi Sankyo, Seagen, Astellas, Ipsen, AstraZeneca, Servier, Bayer, Takeda, Johnson & Johnson, Bristol Myers Squibb, MSD, Amgen, Merck Serono, and Pierre Fabre, and advisory/consultancy fees from Bristol Myers Squibb, MSD, Amgen, Pierre Fabre, Johnson & Johnson, Servier, Bayer, Takeda, Astellas, GSK, Daiichi Sankyo, Pfizer, BeiGene, Jazz Pharmaceuticals, Incyte, Rottapharm Biotech, Merck Serono, Italfarmaco, Gilead Sciences, AstraZeneca, and Agenus outside the submitted work. R. Petty reports grants from Chief Scientist Office (Scotland) and Cancer Research UK during the conduct of the study, as well as grants and personal fees from Amgen, AstraZeneca, Servier, and Astellas, personal fees from Bristol Myers Squibb, and grants from BeiGene, Platinum Discovery, MSD, Moderna, BioNTech, Daiichi Sankyo, and Roche outside the submitted work. No disclosures were reported by the other authors.
Figures

References
-
- Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

