H3K4me2 distinguishes a distinct class of enhancers during the maternal-to-zygotic transition
- PMID: 40638571
- PMCID: PMC12244696
- DOI: 10.1371/journal.pbio.3003239
H3K4me2 distinguishes a distinct class of enhancers during the maternal-to-zygotic transition
Abstract
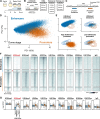
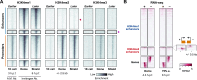
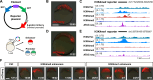
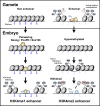
After egg fertilization, an initially silent embryonic genome is transcriptionally activated during the maternal-to-zygotic transition. In zebrafish, maternal vertebrate pluripotency factors Nanog, Pou5f3 (OCT4 homolog), and Sox19b (SOX2 homolog) (NPS) play essential roles in orchestrating embryonic genome activation, acting as "pioneers" that open condensed chromatin and mediate acquisition of activating histone modifications. However, some embryonic gene transcription still occurs in the absence of these factors, suggesting the existence of other mechanisms regulating genome activation. To identify chromatin signatures of these unknown pathways, we profiled the histone modification landscape of zebrafish embryos using CUT&RUN. Our regulatory map revealed two subclasses of enhancers distinguished by presence or absence of H3K4me2. Enhancers lacking H3K4me2 tend to require NPS factors for de novo activation, while enhancers bearing H3K4me2 are epigenetically bookmarked by DNA hypomethylation to recapitulate gamete activity in the embryo, independent of NPS pioneering. Thus, parallel enhancer activation pathways combine to induce transcriptional reprogramming to pluripotency in the early embryo.
Copyright: © 2025 Hurton et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Update of
-
H3K4me2 distinguishes a distinct class of enhancers during the maternal-to-zygotic transition.bioRxiv [Preprint]. 2024 Aug 26:2024.08.26.609713. doi: 10.1101/2024.08.26.609713. bioRxiv. 2024. Update in: PLoS Biol. 2025 Jul 10;23(7):e3003239. doi: 10.1371/journal.pbio.3003239. PMID: 39253505 Free PMC article. Updated. Preprint.
Comment in
-
H3K4me2 marks the enhancer: Enhancer logic in the zebrafish embryo.PLoS Biol. 2025 Jul 11;23(7):e3003257. doi: 10.1371/journal.pbio.3003257. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40644348 Free PMC article.
Similar articles
-
H3K4me2 distinguishes a distinct class of enhancers during the maternal-to-zygotic transition.bioRxiv [Preprint]. 2024 Aug 26:2024.08.26.609713. doi: 10.1101/2024.08.26.609713. bioRxiv. 2024. Update in: PLoS Biol. 2025 Jul 10;23(7):e3003239. doi: 10.1371/journal.pbio.3003239. PMID: 39253505 Free PMC article. Updated. Preprint.
-
Specificity in clustering of gene-specific transcription factors is encoded in the genome.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf625. doi: 10.1093/nar/gkaf625. Nucleic Acids Res. 2025. PMID: 40650976 Free PMC article.
-
Transcriptome Analysis Suggested Striking Transition Around the End of Epiboly in the Gene Regulatory Network Downstream of the Oct4-Type POU Gene in Zebrafish Embryos.Dev Growth Differ. 2025 Jun;67(5):245-269. doi: 10.1111/dgd.70012. Epub 2025 Jun 9. Dev Growth Differ. 2025. PMID: 40490365 Free PMC article.
-
Zygotic Genome Activators, Developmental Timing, and Pluripotency.Curr Top Dev Biol. 2016;116:273-97. doi: 10.1016/bs.ctdb.2015.12.004. Epub 2016 Feb 13. Curr Top Dev Biol. 2016. PMID: 26970624 Review.
-
Nanog Expression in Embryonic Stem Cells - An Ideal Model System to Dissect Enhancer Function.Bioessays. 2017 Dec;39(12):10.1002/bies.201700086. doi: 10.1002/bies.201700086. Epub 2017 Oct 4. Bioessays. 2017. PMID: 28977693 Free PMC article. Review.
Cited by
-
H3K4me2 marks the enhancer: Enhancer logic in the zebrafish embryo.PLoS Biol. 2025 Jul 11;23(7):e3003257. doi: 10.1371/journal.pbio.3003257. eCollection 2025 Jul. PLoS Biol. 2025. PMID: 40644348 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

