Assessment of ex vivo antimalarial drug efficacy in African Plasmodium falciparum parasite isolates, 2016-2023: a genotype-phenotype association study
- PMID: 40639048
- PMCID: PMC12274720
- DOI: 10.1016/j.ebiom.2025.105835
Assessment of ex vivo antimalarial drug efficacy in African Plasmodium falciparum parasite isolates, 2016-2023: a genotype-phenotype association study
Abstract
Background: Given the altered responses to both artemisinins and lumefantrine in Eastern Africa, monitoring antimalarial drug resistance in all African countries is paramount.
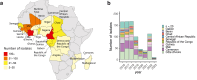
Methods: We measured the susceptibility to six antimalarials using ex vivo growth inhibition assays (IC50) for a total of 805 Plasmodium falciparum isolates obtained from travellers returning to France (2016-2023), mainly from West and Central Africa. Isolates were sequenced using molecular inversion probes (MIPs) targeting forty-three genes across the parasite genome, of which nineteen are drug resistance genes.
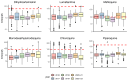
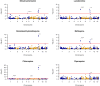
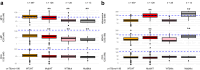
Findings: Ex vivo susceptibility of all assessed antimalarial compounds was consistent with their potent activity. The median IC50 values for the six drugs were 1.1 nM [IQR: 0.8-1.7] for DHA, 16.7 nM [9.9-27.4] for LMF, 29.5 nM [19.1-45.5] for MFQ, 23.4 nM [17.1-39.0] for MDAQ, 26.7 nM [18.0-41.2] for CQ, and 18.5 nM [15.1-24.3] for PPQ. Only four isolates carried a validated pfkelch13 mutation. Multiple mutations in pfcrt and one in pfmdr1 (Asn86Tyr) were significantly associated with altered susceptibility to multiple drugs, and their frequencies decreased over time. Pfcrt and pfmdr1 mutations altered susceptibility to lumefantrine and mefloquine in an additive manner, with the wild-type haplotype (pfcrt K76-pfmdr1 N86) exhibiting the lowest susceptibility.
Interpretation: Our study on P. falciparum isolates from West and Central Africa indicates a low frequency of molecular markers associated with artemisinin resistance and a modest but significant decrease (1.6-2.3X) in the frequency of multidrug resistance markers. These genotypic changes likely mark parasite adaptation to sustained drug pressure and call for intensifying the monitoring of antimalarial drug resistance in Africa.
Funding: This work was supported by the French Ministry of Health (grant to the French National Malaria Reference Centre) and by the Agence Nationale de la Recherche (ANR-17-CE15-0013-03 to JC). JAB was supported by NIH R01AI139520. JR postdoctoral fellowship was funded by Institut de Recherche pour le Développement.
Keywords: Antimalarial drug resistance; Ex-vivo susceptibility; Genotype; Growth inhibition assay.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures

Update of
-
Ex vivo susceptibility to antimalarial drugs and polymorphisms in drug resistance genes of African Plasmodium falciparum, 2016-2023: a genotype-phenotype association study.medRxiv [Preprint]. 2024 Jul 19:2024.07.17.24310448. doi: 10.1101/2024.07.17.24310448. medRxiv. 2024. Update in: EBioMedicine. 2025 Aug;118:105835. doi: 10.1016/j.ebiom.2025.105835. PMID: 39072017 Free PMC article. Updated. Preprint.
References
-
- Conrad M.D., Rosenthal P.J. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis. 2019;19:e338–e351. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

