Structural insights into pre-pore intermediates of alpha-hemolysin in the lipidic environment
- PMID: 40640160
- PMCID: PMC12246456
- DOI: 10.1038/s41467-025-61741-x
Structural insights into pre-pore intermediates of alpha-hemolysin in the lipidic environment
Abstract
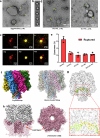
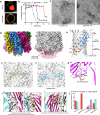
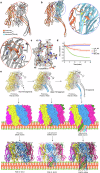
The infectious microbe Staphylococcus aureus releases an array of cytotoxic pore-forming toxins (PFTs) that severely damage the cell membrane during bacterial infection. However, the interaction interfaces between the host cell membrane and toxin were hardly explored. So far, there are no pore, and intermediate structures of these toxins available in the presence of bio-membrane, which could elucidate the pore-forming mechanism. Here, we investigate the structure of different conformational states of this alpha-hemolysin (α-HL/Hla), a β-PFT in lipidic environment using single-particle cryo-EM. Additionally, we explore lipid destabilization by the toxin, using single-molecule imaging, confocal imaging, and validation of lipid-protein interactions using mutational studies. We elucidate eight cryo-EM structures of wildtype α-HL with various liposomes, which are composed of either 10:0 PC or Egg-PC/Cholesterol or Egg-PC/Sphingomyelin or 10:0 PC/Sphingomyelin. Our structural and biophysical studies confirm that different chain lengths and various membrane compositions facilitate the formation of intermediate pre-pores and complete pore species. We also demonstrate that the percentage of pre-pore population increases due to sphingomyelin-induced membrane rigidity. Altogether, this study unveils the structure-function analysis of the pre-pore to pore transition of wildtype α-HL during its crosstalk with the lipid membrane.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Harayama, T. & Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol.19, 281–296 (2018). - PubMed
-
- Dal Peraro, M. & van der Goot, F. G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol14, 77–92 (2016). - PubMed
-
- Bischofberger, M., Iacovache, I. & van der Goot, F. G. Pathogenic pore-forming proteins: function and host response. Cell Host Microbe12, 266–275 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

