Molecular landscape of respiratory infection: A large-scale, multi-centre blood transcriptome dataset
- PMID: 40640204
- PMCID: PMC12246199
- DOI: 10.1038/s41597-025-05488-6
Molecular landscape of respiratory infection: A large-scale, multi-centre blood transcriptome dataset
Abstract
Respiratory infections pose significant challenges to global health, impacting millions of individuals annually. Understanding the molecular mechanisms underlying the pathogenicity of these infections is crucial for developing effective interventions. RNA sequencing provides insights into a patient's global transcriptome changes, facilitating the identification of host gene signatures in response to infection and potential therapeutic targets. Here we present an extensive whole blood transcriptome dataset from a demographically diverse cohort of 502 patients with infections including COVID-19, seasonal coronavirus, influenza A or influenza B, sepsis, septic shock, and co-infections (Viral/Viral, Bacterial/Viral, Bacterial/Viral/Fungal, Viral/Fungal, Viral/ Viral/Fungal). The cohort size and depth of data showcase its potential to unravel respiratory infection pathogenesis for the development of better diagnostics, treatments, and preventive strategies for respiratory infections and future global health crises.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
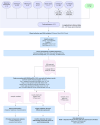
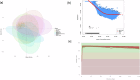
References
-
- Netea, M. G., Schlitzer, A., Placek, K., Joosten, L. A. B. & Schultze, J. L. Innate and Adaptive Immune Memory: an Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe25, 13–26, 10.1016/j.chom.2018.12.006 (2019). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

