Cancer-associated fibroblasts are associated with neo-adjuvant treatment response in oesophageal adenocarcinoma
- PMID: 40640495
- PMCID: PMC12405553
- DOI: 10.1038/s41416-025-03080-8
Cancer-associated fibroblasts are associated with neo-adjuvant treatment response in oesophageal adenocarcinoma
Abstract
Background: Neoadjuvant treatment (NAT) in oesophageal adenocarcinoma (EAC) is characterised by differential responses between patients and treatment modalities. The components of the tumour microenvironment (TME) that contribute to this are unknown. We explored this, focusing on cancer-associated fibroblasts (CAF) an abundant TME component.
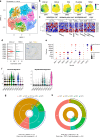
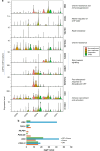
Methods: We performed histopathologic, single-cell RNA sequencing and transcriptomic analysis on 26 patients, stratified by pathological response to NAT, and validated a prognostic model in genomic consortia cohorts. Patient-derived cells were used to model CAF phenotypes in vitro.
Results: We observed changes in the TME in response to the NAT received. Specific changes in fibroblasts correlated with treatment response and altered gene expression associated with NAT type. Three myofibroblastic phenotypes dominate the TME, two of which persist in non-responders and could only be partially re-capitulated in vitro using co-culture with cancer cells or TGF-β. A two-gene NAT fibrotic signature was an independent prognostic indicator in chemo/chemoradiotherapy treated patients (HR = 2.47, p = 0.029).
Conclusions: This study provides a compendium of cell phenotypes in EAC across the current NAT treatment pathway that provides insights into CAF biology and cancer progression. MyoCAFs represent an axis to repurpose agents to enhance current therapies and immunotherapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Hoeppner J, Brunner T, Lordick F, Schmoor C, Kulemann B, Neumann UP, et al. Prospective randomized multicenter phase III trial comparing perioperative chemotherapy (FLOT protocol) to neoadjuvant chemoradiation (CROSS protocol) in patients with adenocarcinoma of the esophagus (ESOPEC trial). J Clin Oncol. 2024;42:LBA1–LBA. - PMC - PubMed
-
- Kelly RJ, Ajani JA, Kuzdzal J, Zander T, Van Cutsem E, Piessen G, et al. Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med. 2021;384:1191–203. - PubMed
-
- Janjigian YY, Van Cutsem E, Muro K, Wainberg Z, Al-Batran SE, Hyung WJ, et al. MATTERHORN: phase III study of durvalumab plus FLOT chemotherapy in resectable gastric/gastroesophageal junction cancer. Future Oncol. 2022;18:2465–73. - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

