Fexofenadine HCl enhances growth, biofilm, and lactic acid production of Limosilactobacillus reuteri and Bifidobacterium longum: implications for allergy treatment
- PMID: 40640705
- PMCID: PMC12247301
- DOI: 10.1186/s12866-025-04130-0
Fexofenadine HCl enhances growth, biofilm, and lactic acid production of Limosilactobacillus reuteri and Bifidobacterium longum: implications for allergy treatment
Abstract
Background: It is evident that various drugs influence the gut microbiota, yet the precise mechanism driving these effects remain ambiguous. Considering the growing recognition of gut microbiota's role in health and disease, it is important to explore how commonly used drugs, such as antihistamines, may alter microbial composition and function. Histamine, an essential interkingdom signaling molecule, shapes bacterial virulence, biofilm formation, and immune regulation. However, the effects of antihistamines on bacterial colonization are mostly unknown. This study aimed to investigate the potential effects of antihistamine exposure on critical factors which affect the pathogenicity and colonization of selected gut bacterial species, such as growth, biofilm formation, and adherence to cell lines, at intestinal concentrations. If antihistamines influence bacterial metabolism or composition, they may consequently affect Short Chain Fatty Acid (SCFA) production, which could have downstream effects on gut homeostasis and immune function. Specifically, we examined the impact of three antihistamines - fexofenadine HCl, cyproheptadine HCl, and desloratadine -on bacteria from the four dominant gut phyla: Bifidobacterium longum, Limosilactobacillus reuteri, Bacteroides fragilis, and Escherichia coli.
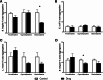
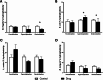
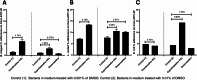
Results: Our results showed that cyproheptadine HCl and desloratadine inhibited the growth of all tested bacteria, whereas fexofenadine HCl promoted the growth of all species except B. longum. Furthermore, cyproheptadine HCl and desloratadine reduced the biofilm-forming capacity of these bacterial species and altered their effects on adherence to Caco-2/HT-29 cell lines aligning with changes in cell surface hydrophobicity: increased cell surface hydrophobicity correlated with greater bacterial adherence to surfaces. In contrast, fexofenadine HCl enhanced biofilm formation and adherence of B. longum and L. reuterii in Caco-2/HT-29 co-cultures. It also led to increased production of lactic and propionic acids, with a statistically significant increase observed in acetic acid levels (p < 0.05).
Conclusion: In summary, our findings suggest that fexofenadine HCl, unlike cyproheptadine HCl and desloratadine, supports the growth, and colonization of probiotic bacteria such as L. reuteri and B. longum with potential anti allergic benefits, and enhancing their SCFA production. Conversely, cyproheptadine HCl and desloratadine suppressed bacterial growth, hinting at potential antimicrobial properties that may warrant exploration for drug repurposing.
Keywords: Bifidobacterium longum; Limosilactobacillus reuteri; Cyproheptadine HCl; Desloratadine; Fexofenadine HCl; Lactic acid.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not Applicable. Consent for publication: Not Applicable. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

