Study on the regulatory mechanism of NsdAsr on rimocidin biosynthesis in Streptomyces rimosus M527
- PMID: 40640785
- PMCID: PMC12243394
- DOI: 10.1186/s12934-025-02784-z
Study on the regulatory mechanism of NsdAsr on rimocidin biosynthesis in Streptomyces rimosus M527
Abstract
Background: We previously identified a regulator NsdAsr, which negatively regulated rimocidin biosynthesis in Streptomyces rimosus M527. However, the exact regulatory mechanism of NsdAsr on rimocidin production remains unknown.
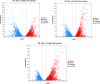
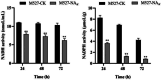
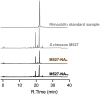
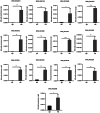
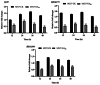
Results: In this study, firstly, transcriptomic data demonstrated that the differentially expressed genes resulting from the over-expression of nsdAsr were primarily associated with several key metabolic pathways, including glycolysis, oxidative phosphorylation, and ribosome-related genes, all of which were downregulated. This directly impacted the concentrations of CoA and NADH, as confirmed by concentration measurement assays. Subsequently, the results of the ChIP-seq experiments revealed that NsdAsr directly binds to 49 target genes. Notably, these include RS18275 and RS18290 (both involved in fatty acid degradation) as well as rpoB (related to DNA transcription). The validity of the ChIP-seq assay for these three genes was further supported by in vitro electrophoretic mobility shift assays. Regarding RS18275 and RS18290, the results revealed that the binding of NsdAsr to these elements led to the downregulation of gene expression. This, in turn, resulted in a decrease in the levels of butyryl-CoA and malonyl-CoA, which are known precursors for rimocidin biosynthesis. Consequently, this negatively impacted on the biosynthesis of rimocidin. In the case of rpoB, the results indicated that NsdAsr binding led to a downregulation of overall protein levels. This was determined by enzymatic activity of report gene GUS and Western blot assay. Consequently, this resulted in a decrease in rimocidin yield.
Conclusion: This study reveals NsdAsr's dual role in limiting rimocidin production by suppressing metabolic precursors and modulating protein expression. Integrated transcriptomic and ChIP-seq analyses provide critical insights into its regulatory mechanisms.
Keywords: Streptomyces rimosus; ChIP-Seq; GUS; NsdAsr; Precursor supply.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
Identification of a gene from Streptomyces rimosus M527 negatively affecting rimocidin biosynthesis and morphological differentiation.Appl Microbiol Biotechnol. 2020 Dec;104(23):10191-10202. doi: 10.1007/s00253-020-10955-8. Epub 2020 Oct 15. Appl Microbiol Biotechnol. 2020. PMID: 33057790
-
Metabolomic analysis of the impact of MtrA on carbon metabolism in Streptomyces coelicolor.Microbiol Spectr. 2025 Aug 5;13(8):e0009625. doi: 10.1128/spectrum.00096-25. Epub 2025 Jun 30. Microbiol Spectr. 2025. PMID: 40586569 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Del Carratore F, Hanko EK, Breitling R, Takano E. Biotechnological application of Streptomyces for the production of clinical drugs and other bioactive molecules. Curr Opin Biotechnol. 2022;77:102762. - PubMed
-
- Cuozzo S, de Moreno de LeBlanc A, LeBlanc JG, Hoffmann N, Tortella GR. Streptomyces genus as a source of probiotics and its potential for its use in health. Microbiol Res. 2023;266:127248. - PubMed
-
- Liu R, Deng Z, Liu T. Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab Eng. 2018;50:74–84. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

