Decoding m6A RNA methylation in kidney disorders: from molecular insights to therapeutic strategies
- PMID: 40640897
- PMCID: PMC12247441
- DOI: 10.1186/s12967-025-06817-4
Decoding m6A RNA methylation in kidney disorders: from molecular insights to therapeutic strategies
Abstract
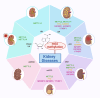
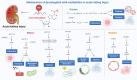
N6-methyladenosine (m6A), the most abundant internal modification in eukaryotic messenger RNA (mRNA) and long noncoding RNA (lncRNA), is dynamically modulated by methyltransferases ("writers"), demethylases ("erasers"), and binding proteins ("readers"). As a central epitranscriptomic regulator, m6A governs RNA stability, splicing, translation, and degradation, thereby orchestrating a wide range of physiological and pathological pathways. Accumulating evidence has underscored its pivotal involvement in the pathogenesis of kidney disorders. This review delineates the regulatory landscape of m6A methylation across various kidney diseases, with emphasis on diabetic nephropathy (DN), acute kidney injury (AKI), chronic kidney disease (CKD), focal segmental glomerulosclerosis (FSGS), lupus nephritis (LN), hyperuricemic nephropathy (HN), autosomal dominant polycystic kidney disease (ADPKD), and clear cell renal cell carcinoma (ccRCC). Disease-specific alterations in m6A levels and the expression patterns of core regulators, including METTL3, METTL14, FTO, ALKBH5, YTH domain proteins, and IGF2BPs, are systematically summarized. By elucidating their roles in inflammation, fibrosis, apoptosis, and metabolic imbalance, this review highlights the translational potential of m6A-centric interventions and offers novel insights into epitranscriptomic regulation within renal pathophysiology.
Keywords: Clinical significance; Expression profiles; Kidney diseases; Regulatory mechanisms; m6A modification.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All the authors have read and approved the final manuscript. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Unlocking the potential: m6A-RNA methylation in severe epidermolysis bullosa simplex.Biosci Rep. 2025 Jul 22;45(7):BSR20253141. doi: 10.1042/BSR20253141. Biosci Rep. 2025. PMID: 40700032
-
METTL3-Mediated N 6 -Methyladenosine mRNA Modification and cGAS-STING Pathway Activity in Kidney Fibrosis.J Am Soc Nephrol. 2024 Oct 1;35(10):1312-1329. doi: 10.1681/ASN.0000000000000428. Epub 2024 Jun 10. J Am Soc Nephrol. 2024. PMID: 39352860
-
Novel landscapes of N6-methyladenosine modification of mitochondrial oxidative stress in organ fibrosis.Eur J Pharmacol. 2025 Sep 15;1003:177888. doi: 10.1016/j.ejphar.2025.177888. Epub 2025 Jul 1. Eur J Pharmacol. 2025. PMID: 40609613 Review.
-
Comprehensive genomic characterization and differential expression analysis of m6A RNA methylation regulatory proteins (writers, readers, and erasers) in Suaeda Salsa under abiotic stress condition.BMC Plant Biol. 2025 Jul 31;25(1):996. doi: 10.1186/s12870-025-07060-1. BMC Plant Biol. 2025. PMID: 40745283 Free PMC article.
-
RNA N 6-Methyladenosine Modification in Normal and Malignant Hematopoiesis.Adv Exp Med Biol. 2019;1143:75-93. doi: 10.1007/978-981-13-7342-8_4. Adv Exp Med Biol. 2019. PMID: 31338816 Review.
References
-
- Li B, Qu L, Yang J, RNA-Guided RNA, Modifications. Biogenesis, functions, and applications. Acc Chem Res. 2023;56:3198–210. 10.1021/acs.accounts.3c00474. - PubMed
-
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17:365–72. 10.1038/nrg.2016.47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82360157/National Natural Science Foundation of China
- U21A20350/National Natural Science Foundation of China
- 2023C04011/Key Research and Development Program of Science and Technology Department of Zhejiang Province
- 2023AB018-03/Key Research and Development Project of Xinjiang Production and Construction Corps Science and Technology Bureau
- Y202147593/Scientific Research Fund of Zhejiang Provincial Education Department
LinkOut - more resources
Full Text Sources
Medical
Research Materials

