Fatty acid synthase-derived lipid stores support breast cancer metastasis
- PMID: 40640944
- PMCID: PMC12247306
- DOI: 10.1186/s40170-025-00404-3
Fatty acid synthase-derived lipid stores support breast cancer metastasis
Abstract
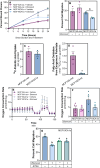
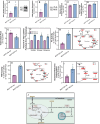
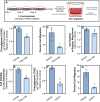
Lipid accumulation is associated with breast cancer metastasis. However, the mechanisms underlying how breast cancer cells increase lipid stores and their functional role in disease progression remain incompletely understood. Herein we quantified changes in lipid metabolism and characterized cytoplasmic lipid droplets in metastatic versus non-metastatic breast cancer cells. 14C-labeled palmitate was used to determine differences in fatty acid (FA) uptake and oxidation. Despite similar levels of palmitate uptake, metastatic cells increase lipid accumulation and oxidation of endogenous FAs compared to non-metastatic cells. Isotope tracing also demonstrated that metastatic cells support increased de novo lipogenesis by converting higher levels of glutamine and glucose into the FA precursor, citrate. Consistent with this, metastatic cells displayed increased levels of fatty acid synthase (FASN) and de novo lipogenesis. Genetic depletion or pharmacologic inhibition of FASN reduced cell migration, survival in anoikis assays, and in vivo metastasis. Finally, global proteomic analysis indicated that proteins involved in proteasome function, mitotic cell cycle, and intracellular protein transport were reduced following FASN inhibition of metastatic cells. Overall, these studies demonstrate that breast cancer metastases accumulate FAs by increasingde novo lipogenesis, storing TAG as cytoplasmic lipid droplets, and catabolizing these stores to drive several FAO-dependent steps in metastasis.
Keywords: Breast cancer; FASN; Fatty acid synthase; Fatty acids; Lipid droplet; Lipid metabolism; Lipid storage; Mass spectrometry; Metastasis; TNBC.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





Update of
-
Fatty Acid Synthase-Derived Lipid Stores Support Breast Cancer Metastasis.Res Sq [Preprint]. 2024 Dec 5:rs.3.rs-5510550. doi: 10.21203/rs.3.rs-5510550/v1. Res Sq. 2024. Update in: Cancer Metab. 2025 Jul 10;13(1):35. doi: 10.1186/s40170-025-00404-3. PMID: 39678343 Free PMC article. Updated. Preprint.
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9(5):311–20. - PubMed
-
- Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

