Identification of serum exosome proteins in systemic sclerosis with interstitial lung disease by aptamer proteomics
- PMID: 40640963
- PMCID: PMC12243147
- DOI: 10.1186/s13075-025-03595-8
Identification of serum exosome proteins in systemic sclerosis with interstitial lung disease by aptamer proteomics
Abstract
Objective: A major unmet need for Systemic Sclerosis (SSc) clinical management is the absence of well validated biomarkers for early diagnosis of SSc-associated interstitial lung disease (SSc-ILD). The objective of this study was to identify proteins contained within serum exosomes that may serve as potential biomarkers to differentiate patients with Diffuse SSc without SSc-ILD from patients with Diffuse SSc with SSc-ILD employing aptamer-based proteomics.
Methods: Serum exosomes were isolated from two cohorts of patients. The first cohort included 15 patients with Diffuse SSc without SSc-ILD and 14 patients with Diffuse SSc with SSc-ILD and the second cohort included 12 patients with Diffuse SSc with SSc-ILD and 12 patients with Diffuse SSc without SSc-ILD. SOMAscan aptamer proteomics was performed with the first cohort and quantified the concentration levels of 1,305 proteins. Significant associations of differentially elevated or reduced proteins (p < 0.05 |FC|>1.2) discriminating between the two SSc clinical subsets were assessed. Validation of the results obtained from the proteomics analysis of the first cohort was performed with the second cohort.
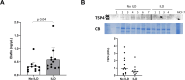
Results: The aptamer proteomic analysis identified 29 proteins increased and 9 proteins decreased in SSc with SSc-ILD as compared to SSc without SSc-ILD. Principal component analysis using the 20 most significantly differentially expressed proteins resulted in excellent separation of the two SSc clinical subsets. Most of the differentially increased proteins converged around enhanced inflammatory responses, immune cell activation, cell death, and abnormal vascular functions and several of them displayed a highly significant correlation with the CO Diffusion Capacity Levels.
Conclusion: Aptamer proteomic analysis of circulating exosomes from patients with Diffuse SSc with and without SSc-ILD identified several biologically plausible biomarkers that may be of value to differentiate these two SSc clinical subsets.
Keywords: Aptamer proteomics; Interstitial lung disease; Serum exosomes; Systemic sclerosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All authors have read and agreed to its content and are accountable for all aspects of the accuracy and integrity of the manuscript in accordance with ICMJE criteria. Consent for publication: This article is original and has not been published or currently under consideration by another journal. The authors agree to the terms of the BioMed Central Copyright and License Agreement, Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
An exploratory analysis of differences in serum protein expression by sex in patients with systemic sclerosis associated interstitial lung disease.BMC Pulm Med. 2025 Jan 13;25(1):16. doi: 10.1186/s12890-024-03474-z. BMC Pulm Med. 2025. PMID: 39806325 Free PMC article.
-
Dynamics of interstitial lung disease following immunosuppressive treatment differ between antisynthetase syndrome and systemic sclerosis.Ther Adv Respir Dis. 2025 Jan-Dec;19:17534666251336896. doi: 10.1177/17534666251336896. Epub 2025 May 8. Ther Adv Respir Dis. 2025. PMID: 40337907 Free PMC article.
-
Assessing the MUC5B promoter variant in a large cohort of systemic sclerosis-associated interstitial lung disease.RMD Open. 2025 Aug 12;11(3):e005754. doi: 10.1136/rmdopen-2025-005754. RMD Open. 2025. PMID: 40803819 Free PMC article.
-
Cyclophosphamide for connective tissue disease-associated interstitial lung disease.Cochrane Database Syst Rev. 2018 Jan 3;1(1):CD010908. doi: 10.1002/14651858.CD010908.pub2. Cochrane Database Syst Rev. 2018. PMID: 29297205 Free PMC article.
-
The role of lung biopsy for diagnosis and prognosis of interstitial lung disease in systemic sclerosis: a systematic literature review.Respir Res. 2024 Mar 23;25(1):138. doi: 10.1186/s12931-024-02725-1. Respir Res. 2024. PMID: 38521926 Free PMC article.
References
-
- Gabrielli A, Avvedimento EV, Krieg T, Scleroderma. N Engl J Med. 2009;360:1989–2003. - PubMed
-
- Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, Varga J. Systemic sclerosis. Nat Rev Dis Prim. 2015;1:15002. - PubMed
-
- Denton CP, Khanna D. Systemic sclerosis. Lancet. 2017;390:1685–99. - PubMed
-
- Asano Y. Systemic sclerosis. J Dermatol. 2018;45:128–38. - PubMed
-
- Katsumoto TR, Whitfield ML, Connolly MK. The pathogenesis of systemic sclerosis. Annu Rev Pathol. 2011;28:509–37. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

