Clonorchis sinensis infection remodels chromatin accessibility in hepatocellular carcinoma
- PMID: 40640964
- PMCID: PMC12247385
- DOI: 10.1186/s13071-025-06909-6
Clonorchis sinensis infection remodels chromatin accessibility in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is a major global health concern, accounting for a significant proportion of liver cancer cases and related deaths. Clonorchis sinensis (C. sinensis) infection, a recognized carcinogen, has been implicated in the progression of liver diseases, including HCC. However, the precise epigenetic mechanisms underlying C. sinensis-associated HCC remain to be elucidated.
Methods: To investigate the role of chromatin accessibility in C. sinensis-related HCC progression, we performed an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and RNA sequencing (RNA-seq) analyses of C. sinensis-infected (C. sinensis+) and non-C. sinensis-infected (C. sinensis-) HCC tumors. Integrated analyses were conducted to assess chromatin accessibility, transcription factor (TF) motifs, and histone modifications using ATAC-seq, RNA-seq, and classical chromatin immunoprecipitation-sequencing (ChIP-seq) datasets. A scratch wound assay was used to evaluate the effects of C. sinensis excretory/secretory products (CsESPs) on HCC cell migration.
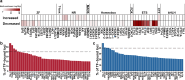
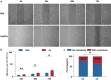
Results: ATAC-seq analysis revealed 9,396 differentially accessible regions (DARs) in C. sinensis+ HCC tumors compared with C. sinensis- HCC tumors. Additionally, several crucial TFs enriched in DARs were identified, including HNF4A, FOXO1, ELF4, and RELA. Combined ATAC-seq and RNA-seq analyses further revealed differentially expressed genes (DEGs) associated with metabolism, immune regulation, and cytoskeletal dynamics. Chromatin accessibility was closely associated with histone modifications such as H3K9ac, H3K4me2, H3K4me3, H3K27ac, H3K4me1, and CTCF binding. Notably, C. sinensis infection significantly increased the migratory capacity of HCC cells, as confirmed by molecular assays and clinical observations.
Conclusions: Our study demonstrates that C. sinensis infection remodels chromatin accessibility and may contribute to HCC progression. Our work offers valuable insights into the pathogenesis of HCC in the context of parasitic infection and lays the groundwork for future biomarker and therapeutic target discovery.
Keywords: Clonorchis sinensis; Chromatin accessibility; Hepatocellular carcinoma; Transcription factor; Tumor progression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study received approval from the Ethics Committee of the Affiliated Cancer Hospital of Guangxi Medical University (LW. 2024093) and adhered to the ethical guidelines set forth in the Helsinki Declaration of 1964. To safeguard patient confidentiality, individuals’ identities in this study were anonymized using computer-generated ID numbers. Upon admission, all patients provided written consent for the analysis and publication of their anonymized medical data for research purposes. Consent for publication: All authors have approved the manuscript for submission. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Multi-omics analysis of lactate metabolism gene regulation in Clonorchis sinensis-associated hepatocellular carcinoma.Parasit Vectors. 2025 Jul 27;18(1):301. doi: 10.1186/s13071-025-06947-0. Parasit Vectors. 2025. PMID: 40717080 Free PMC article.
-
Exploring the mechanism of Naringenin in the treatment of hepatocellular carcinoma based on mRNA sequencing and experimental validation.Sci Rep. 2025 Jul 2;15(1):23109. doi: 10.1038/s41598-025-09013-y. Sci Rep. 2025. PMID: 40593127 Free PMC article.
-
ARID1A deficiency promotes malignant proliferation of hepatocellular carcinoma by activating HDAC7/ENO1 signaling pathway.Hepatol Commun. 2025 Jun 19;9(7):e0738. doi: 10.1097/HC9.0000000000000738. eCollection 2025 Jul 1. Hepatol Commun. 2025. PMID: 40536557 Free PMC article.
-
Prevalence estimates of Opisthorchis viverrini and Clonorchis sinensis infection in the Greater Mekong subregion: a systematic review and meta-analysis.Infect Dis Poverty. 2024 May 8;13(1):33. doi: 10.1186/s40249-024-01201-8. Infect Dis Poverty. 2024. PMID: 38720371 Free PMC article.
-
Percutaneous ethanol injection or percutaneous acetic acid injection for early hepatocellular carcinoma.Cochrane Database Syst Rev. 2015 Jan 26;1(1):CD006745. doi: 10.1002/14651858.CD006745.pub3. Cochrane Database Syst Rev. 2015. PMID: 25620061 Free PMC article.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2024;74:229–63. 10.3322/caac.21834. - PubMed
-
- Hepatocellular carcinoma. Nature reviews Disease primers. 2021;7:7; 10.1038/s41572-021-00245-6. - PubMed
-
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10:321–2. 10.1016/s1470-2045(09)70096-8. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

