Methanotrophic Flexibility of 'Ca. Methanoperedens' and Its Interactions With Sulphate-Reducing Bacteria in the Sediment of Meromictic Lake Cadagno
- PMID: 40641180
- PMCID: PMC12246660
- DOI: 10.1111/1462-2920.70133
Methanotrophic Flexibility of 'Ca. Methanoperedens' and Its Interactions With Sulphate-Reducing Bacteria in the Sediment of Meromictic Lake Cadagno
Abstract
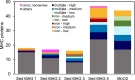
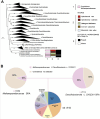
The greenhouse gas methane is an important contributor to global warming, with freshwater sediments representing important potential methane sources. Anaerobic methane-oxidising archaea mitigate methane release into the atmosphere by coupling the oxidation of methane to the reduction of extracellular electron acceptors or through interspecies electron transfer with microbial partners. Understanding their metabolic flexibility and microbial interactions is crucial to assess their role in global methane cycling. Here, we investigated anoxic sediments of the meromictic freshwater Lake Cadagno (Switzerland), where 'Ca. Methanoperedens' co-occur with a specific sulphate-reducing bacterium, with metagenomics and long-term incubations. Incubations were performed with different electron acceptors, revealing that manganese oxides supported highest CH4 oxidation potential but enriched for 'Ca. Methanoperedens' phylotypes that were hardly present in the inoculum. Combining data from the inoculum and incubations, we obtained five 'Ca. Methanoperedens' genomes, each harbouring different extracellular electron transfer pathways. In a reconstructed Desulfobacterota QYQD01 genome, we observed large multi-heme cytochromes, type IV pili, and a putative loss of hydrogenases, suggesting facultative syntrophic interactions with 'Ca. Methanoperedens'. This research deepens our understanding of the metabolic flexibility and potential interspecific interactions of 'Ca. Methanoperedens' in freshwater lakes.
© 2025 The Author(s). Environmental Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Alneberg, J. , Bjarnason B. S., de Bruijn I., et al. 2014. “Binning Metagenomic Contigs by Coverage and Composition.” Nature Methods 11: 1144–1146. - PubMed
-
- Arshad, A. , Dalcin Martins P., Frank J., Jetten M. S. M., Op den Camp H. J. M., and Welte C. U.. 2017. “Mimicking Microbial Interactions Under Nitrate‐Reducing Conditions in an Anoxic Bioreactor: Enrichment of Novel Nitrospirae Bacteria Distantly Related to Thermodesulfovibrio .” Environmental Microbiology 19: 4965–4977. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

