Experimental study on the role and biomarker potential of CX3CR1 in osteoarthritis
- PMID: 40641371
- PMCID: PMC12258232
- DOI: 10.1080/07853890.2025.2529577
Experimental study on the role and biomarker potential of CX3CR1 in osteoarthritis
Abstract
Background: Osteoarthritis (OA) is a chronic joint disorder marked by progressive degeneration of articular cartilage and the formation of secondary osteophytes. Despite extensive research, the underlying molecular mechanisms remain poorly understood. This study aimed to identify OA-associated genes and elucidate the molecular pathways implicated, with the goal of discovering reliable diagnostic biomarkers.
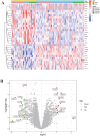
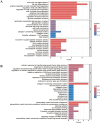
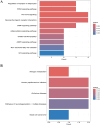
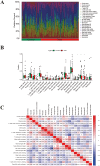
Methods: The microarray dataset was retrieved from the Gene Expression Omnibus (GEO) and analyzed using R software to identify the signature gene, CX3CR1. Differentially expressed genes (DEGs) correlated with CX3CR1 were subsequently subjected to Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and immune infiltration analyses. A ceRNA regulatory network was also constructed. Vali-dation of CX3CR1 expression was conducted through qRT-PCR, Western blotting, and immunohistochemistry.
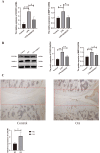
Results: CX3CR1 emerged as a candidate gene significantly associated with OA, exhibiting regulatory roles primarily in lipid metabolism-related and extra-cellular matrix-related biological processes and signaling cascades. The infiltration levels of immune cells, particularly activated mast cells, appeared to modulate OA progression. Both in vitro and in vivo experiments demonstrated elevated CX3CR1 expression in OA tissues relative to controls, with a robust positive correlation observed between CX3CR1 and MMP13 levels.
Conclusion: CX3CR1 represents a potential biomarker for OA diagnosis and therapeutic targeting, exerting its effects by modulating lipid metabolism, extracellular matrix dynamics, and immune cell infiltration.
Keywords: Bioinformatic; CX3CR1; experiment; extracellular matrix; lipid metabolism; osteoarthritis; signature gene.
Conflict of interest statement
No commercial or financial relationships were identified during the study that could be interpreted as a potential conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
