Genetic diversity of Bordetella bronchiseptica isolates obtained from primates
- PMID: 40641881
- PMCID: PMC12243275
- DOI: 10.3389/fmicb.2025.1571660
Genetic diversity of Bordetella bronchiseptica isolates obtained from primates
Abstract
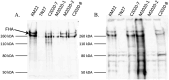
Bordetella bronchiseptica is a highly contagious bacterial respiratory pathogen with a broad host range of wild and domesticated mammals that can cause a variety of clinical disease outcomes ranging from asymptomatic carriage to severe pneumonia. The goal of this study was to evaluate the genetic diversity of B. bronchiseptica isolates obtained from primates and evaluate the antimicrobial resistance harbored by these isolates. Two isolates were identified as belonging to B. bronchiseptica lineage II and 13 isolates represented new sequence types within B. bronchiseptica lineage I clonal complex 6. The lineage II isolates harbored the lowest sequence identity observed across all genes evaluated and did not contain several well characterized virulence and fimbrial genes. Western blotting revealed no reactivity to a lineage II strain when using antibodies generated against pertactin (PRN) from a lineage I-1 strain or antibodies generated against a domain of filamentous haemagglutinin (FHA) from a lineage I-1 strain. Isolates harbored variation within the wbm locus containing genes encoding for the expression of antigenically distinct O-antigen types and the cya operon was replaced by the ptp operon in several isolates, expanding the phylogenetic distribution of this operon replacement. Thirteen isolates exhibited phenotypic resistance to four antibiotic classes tested, however the Bordetella-specific β-lactamase was the only antimicrobial resistance gene identified. Collectively, the data in this report expands the known phylogenetic diversity and genetic variation of B. bronchiseptica isolates.
Keywords: Bordetella bronchiseptica; antimicrobial resistance (AMR); comparative genomics; primate; virulence factor; whole-genome sequencing (WGS).
Copyright © 2025 Nicholson, Shore, Wang, Zimmerman and Merkel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










References
LinkOut - more resources
Full Text Sources

