i-DENV: development of QSAR based regression models for predicting inhibitors targeting non-structural (NS) proteins of dengue virus
- PMID: 40642016
- PMCID: PMC12241036
- DOI: 10.3389/fphar.2025.1605722
i-DENV: development of QSAR based regression models for predicting inhibitors targeting non-structural (NS) proteins of dengue virus
Abstract
Introduction: Dengue virus (DENV) is a significant global arboviral threat with fatal potential, currently lacking effective antiviral treatments or a universally applicable vaccine. In response to this unmet need, we developed the "i-DENV" web server to facilitate structure-based drug prediction targeting key viral proteins.
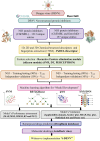
Methods: The i-DENV platform focuses on the NS3 protease and NS5 polymerase of DENV using machine learning techniques (MLTs) and quantitative structure-activity relationship (QSAR) modeling. A total of 1213 and 157 unique compounds, along with their IC50 values targeting NS3 and NS5 respectively, were retrieved from the ChEMBL and DenvInD databases. Molecular descriptors and fingerprints were computed and used to train multiple regression-based MLTs, including SVM, RF, kNN, ANN, XGBoost, and DNN, with ten-fold cross-validation.
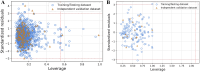
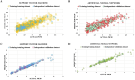
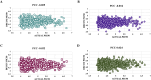
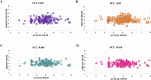
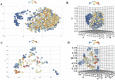
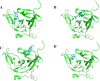
Results: The best-performing SVM and ANN models achieved Pearson correlation coefficients (PCCs) of 0.857/0.862 (NS3) and 0.982/0.964 (NS5) on training/testing sets, and 0.870/0.894 (NS3) and 0.970/0.977 (NS5) on independent validation sets. Model robustness was supported through scatter plots, chemical clustering, statistical analyses, decoy set etc. Virtual screening identified Micafungin, Oritavancin, and Iodixanol as top hits for NS2B/NS3 protease, and Cangrelor, Eravacycline, and Baloxavir marboxil for NS5 polymerase. Molecular docking further confirmed strong binding affinities of these compounds.
Discussion: Our in-silico findings suggest these repurposed drugs as promising antiviral candidates against DENV. However, further in vitro and in vivo studies are essential to validate their therapeutic potential. The i-DENV web server is freely accessible at http://bioinfo.imtech.res.in/manojk/idenv/, offering a structure-specific drug prediction platform for DENV research and antiviral drug discovery.
Keywords: QSAR; algorithm; antivirals; artificial intelligence; machine learning; web server.
Copyright © 2025 Gautam, Thakur and Kumar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures





References
-
- Aguilera-Pesantes D., Robayo L. E., Méndez P. E., Mollocana D., Marrero-Ponce Y., Torres F. J., et al. (2017). Discovering key residues of dengue virus NS2b-NS3-protease: new binding sites for antiviral inhibitors design. Biochem. Biophys. Res. Commun. 492 (4), 631–642. 10.1016/j.bbrc.2017.03.107 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

