The complete chloroplast genome of Cyrtosia nana (Rolfe ex Downie) Garay and its phylogenetic analysis
- PMID: 40642020
- PMCID: PMC12243006
- DOI: 10.1080/23802359.2025.2528572
The complete chloroplast genome of Cyrtosia nana (Rolfe ex Downie) Garay and its phylogenetic analysis
Abstract
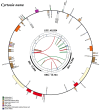
Cyrtosia nana is a mycotrophic orchid distributed in southwestern China and Thailand. Here, we sequenced and assembled its complete chloroplast (cp) genome using next-generation sequencing (NGS) and established a robust phylogenetic framework, providing a foundation for further studies on evolution of related taxa. The complete chloroplast genome was 85,766 bp, including a pair of inverted repeat regions (12,999 bp), a small single-copy region (14,744 bp), and a large single-copy region (45,024 bp). It encodes 47 unique genes, including 28 protein-coding genes, 16 tRNA genes, and 3 rRNA genes. Phylogenetic analysis revealed that C. nana was closely related to C. lindleyana and C. septentrionalis.
Keywords: Cyrtosia nana; chloroplast genome; mycotrophic; phylogeny.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


Similar articles
-
Plastome and transcriptome analysis reveals a degenerated photosynthesis-related pathway in the mycoheterotrophic vanilloid orchid Cyrtosia lindleyana.BMC Plant Biol. 2025 Jul 2;25(1):841. doi: 10.1186/s12870-025-06850-x. BMC Plant Biol. 2025. PMID: 40604445 Free PMC article.
-
Complete chloroplast genome of two Brachycorythis (Orchidaceae) species from China: comparative analysis and phylogenetic implications.BMC Genomics. 2025 Jul 2;26(1):632. doi: 10.1186/s12864-025-11791-8. BMC Genomics. 2025. PMID: 40604395 Free PMC article.
-
Comparison of Chloroplast Genome Sequences of Saxifraga umbellulata var. pectinata in Qinghai-Xizang Plateau.Genes (Basel). 2025 Jun 30;16(7):789. doi: 10.3390/genes16070789. Genes (Basel). 2025. PMID: 40725445 Free PMC article.
-
Incentives for preventing smoking in children and adolescents.Cochrane Database Syst Rev. 2017 Jun 6;6(6):CD008645. doi: 10.1002/14651858.CD008645.pub3. Cochrane Database Syst Rev. 2017. PMID: 28585288 Free PMC article.
-
Ablative and non-surgical therapies for early and very early hepatocellular carcinoma: a systematic review and network meta-analysis.Health Technol Assess. 2023 Dec;27(29):1-172. doi: 10.3310/GK5221. Health Technol Assess. 2023. PMID: 38149643 Free PMC article.
References
-
- Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C.. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. doi: 10.1093/molbev/msu252. - DOI - PubMed
-
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Tsi ZH, Lou YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China, vol. 25. Beijing: Science Press, p. 169.
-
- Divakaran M, Rafieah NTF.. 2021. Genetic resources of the universal flavor, Vanilla. In Herbs and spices (new processing technologies). 13. Books on Demand GmbH, Hamburg. doi: 10.5772/intechopen.99043. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
