Amplification of select autonomous HERV loci and surrounding host gene transcription in monocytes from patients with post-acute sequelae of COVID-19
- PMID: 40642078
- PMCID: PMC12241865
- DOI: 10.3389/fimmu.2025.1621657
Amplification of select autonomous HERV loci and surrounding host gene transcription in monocytes from patients with post-acute sequelae of COVID-19
Abstract
Background: The human genome contains approximately 3,200 near full-length autonomous human endogenous retroviral (HERV) genomes distributed across the 23 chromosomes. These autonomous HERV proviral genomes include long terminal repeats (LTRs) capable of promoting RNA transcription. In quiescent cells, most HERV loci remain transcriptionally silent. However, environmental changes, such as epigenetic remodeling of chromatin, can activate these silenced loci.
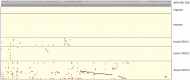
Methods: To study HERV reactivation, we previously analyzed autonomous HERV expression patterns in monocytes isolated from peripheral blood mononuclear cells (PBMCs) identified in single-cell RNA sequencing (scRNA-seq) databases using the Azimuth application. We developed a Window-based HERV Alignment (WHA) method, which analyzes aligned DNA sequences using sequential, non-overlapping windows of defined lengths. Samples were scored as positive (>= 9 good/usable windows) or negative (<= 8 good/usable windows).
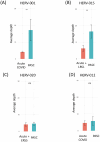
Results: Using WHA, we established a control set from 31 normal individuals, with fewer than 8 windows at selected HERV loci. We analyzed scRNA-seq data from three studies of hospitalized COVID-19 patients and found distinct HERV expression patterns in monocytes. Unique patterns were also found in patients with influenza, Dengue virus, or sepsis. We next examined HERV expression at early (<7 days) and late (>14 days) timepoints post COVID-19 recovery and detected HERV loci in both groups. Analyzing 12 patients with post-acute sequelae of COVID-19 (PASC), we identified three HERV loci expressed in all patients. Some loci showed amplified numbers of good/usable windows, indicating longer transcripts and greater sequence depth. The most amplified locus was located within an intron of JAKMIP2, which, along with neighboring host genes, also showed increased transcription.
Conclusion: Previous studies have shown that viral infections, including COVID-19, influenza, and Dengue virus, as well as sepsis, can induce innate immune memory in monocytes through epigenetic remodeling of hematopoietic stem and myeloid precursor cells. The identification of co-amplified HERV loci and neighboring host gene transcripts in monocytes from PASC patients suggests expansion of epigenetically remodeled myeloid progenitors. The identification of these HERV-host gene patterns provides a foundation needed to understand the clinical features of patients with PASC.
Keywords: ScRNA-seq; epigenetic remodeling; human endogenous retrovirus (HERV); monocytes; post-acute COVID sequelae.
Copyright © 2025 Koo and Morrow.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

