Engineering functionally-optimized aptamers against SARS-Cov-2 for blocking spike-ACE2 interaction and aptasensor detection
- PMID: 40642289
- PMCID: PMC12241384
- DOI: 10.1016/j.mtbio.2025.102020
Engineering functionally-optimized aptamers against SARS-Cov-2 for blocking spike-ACE2 interaction and aptasensor detection
Abstract
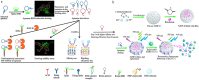
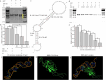
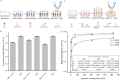
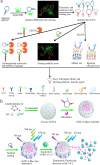
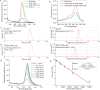
Both the limited research about structure-function relationship and the ill-defined process of conformational dynamic change greatly impede the development of aptamer engineering transformation and seriously restrict the practical applications of aptamers. In this work, an optimization strategy combining exonuclease III (Exo III) digestion and in silico simulation was presented for the first time for constructing high-affinity and functional aptamers and clarifying the three-dimensional (3D) structure of aptamer-target complexes and the conformational dynamic conversion in the process of aptamer recognizing its target. As a demonstration, the parent aptamer (Apt2) against SARS-CoV-2 spike subunit 1 (S1) was mutated or truncated at the predicted binding sites to produce eight derivatives (Seq1-Seq8). The progeny Seq3 exhibited a higher affinity for S1 and a better blocking effect on S1-angiotensin-converting enzyme 2 (ACE2) interaction compared to Apt2. Subsequently, Seq3 sealed the pores of nickel-doped zeolitic imidazolate framework-8 (NZIF-8) loaded with Rhodanine (Rho) to fabricate the aptasensor (NZIF-8-Rho-Apt) for inactivated virus detection, showing excellent performances in spiked actual samples. Therefore, this post systematic evolution of ligands by exponential enrichment (post-SELEX) is a very effective and general strategy for acquiring functionally-optimized aptamers.
Keywords: Aptamer optimization; Aptasensors; Exonuclease III digestion; In silico simulation; Inactivated virus detection; SARS-CoV-2 spike.
© 2025 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Geng L., Liu J., Zhang W., Wang H., Huang J., Wang G., Hu M., Dong H., Sun J., Fang M., Guo Y., Sun X. Preparation of dual recognition adsorbents based on molecularly imprinted polymers and aptamer for highly sensitive recognition and enrichment of ochratoxin A. J. Hazard. Mater. 2024;476 doi: 10.1016/j.jhazmat.2024.135112. - DOI - PubMed
-
- Lai X., Zhang X., Lai J., Zhao W., Song Z., Chen Y., Ud din M., Munawer M.F., Jiang H., Liu X., Wang X. Targeted self-assembled anti-NFκB AuNCs-aptamer nanoplatform for precise theranostics via tailored follicle regeneration. Mater. Today Bio. 2025;32 doi: 10.1016/j.mtbio.2025.101774. - DOI - PMC - PubMed
-
- Lai X., Liu T., Guo Z., Wang Y., Xiao J., Xia Q., Liu X., Jiang H., Wang X. In situ formed fluorescent gold nanoclusters inhibit hair follicle regeneration in oxidative stress microenvironment via suppressing NFκB signal pathway. Chin. Chem. Lett. 2025;36 doi: 10.1016/j.cclet.2024.109762. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

