Characterization of a novel Phietavirus genus bacteriophage and its potential for efficient transfer of modified shuttle plasmids to Staphylococcus aureus strains of different clonal complexes
- PMID: 40642983
- PMCID: PMC12323379
- DOI: 10.1128/spectrum.03332-24
Characterization of a novel Phietavirus genus bacteriophage and its potential for efficient transfer of modified shuttle plasmids to Staphylococcus aureus strains of different clonal complexes
Abstract
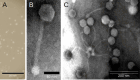
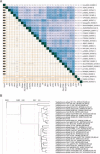
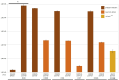
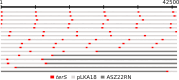
Staphylococcus aureus is a significant human pathogen responsible for various nosocomial and community-acquired infections, leading to considerable morbidity and mortality worldwide. Temperate bacteriophages contribute to its virulence and facilitate the dissemination of pathogenicity traits. We isolated a novel siphovirus of the Phietavirus genus, ASZ22RN, derived from a prophage of an S. aureus clonal complex 7 strain and capable of propagating in the prophage-free laboratory strain RN4220. ASZ22RN either productively infected or lysed from without all 47 tested S. aureus clinical strains across 12 clonal complexes (CCs), demonstrating its ability to puncture their cell envelopes. When ASZ22RN was propagated in RN4220 cells harboring an S. aureus-Escherichia coli plasmid replicating via theta mode, it transduced the plasmid to plasmid-free RN4220 with low frequency. The transduction frequency increased by nearly five orders of magnitude when the plasmid contained a fragment of ASZ22RN DNA (terS). Most terS+ plasmid-transducing particles carried plasmid concatamers, while some carried plasmid-phage DNA hybrids, as demonstrated by DNA sequencing. Strains from all tested CCs served as recipients for transduction, regardless of the presence of type I restriction-modification enzymes targeting plasmid/phage DNA, or prophages with lysis-lysogeny switch regions conferring superinfection immunity to ASZ22RN. Our results indicate that intracellular phage defense systems do not prevent phage-mediated plasmid transfer and demonstrate a simple method for introducing plasmids constructed in E. coli into clinical S. aureus isolates. Moreover, the presence of the ASZ22RN lysis-lysogeny switch region in 21% of tested ASZ22RN-resistant strains highlights superinfection exclusion as a dominant mechanism of resistance to siphoviruses in staphylococci.
Importance: This study highlights the capacity of a newly isolated staphylococcal Phietavirus, ASZ22RN, to transfer a low-copy-number shuttle Staphylococcus aureus-Escherichia coli plasmid to various S. aureus strains representing major clonal complexes from among clinical isolates. By increasing the plasmid transduction efficiency in an ASZ22RN-specific manner, we show that the primary factor determining a given strain's ability to be a recipient in transduction is the capacity of transducing phage to puncture the cell envelopes of this strain. This can be determined not only based on productive phage infection but also lysis from without. Major intracellular mechanisms protecting S. aureus from productive phage infection do not impede the transduction-mediated acquisition of plasmids. Moreover, the lack of phage DNA in most of the plasmid-transducing virions indicates the lack of phage contamination in most transductants. Our results offer a promising approach for developing efficient pipelines to introduce plasmids constructed in E. coli to clinical S. aureus isolates.
Keywords: Phietavirus; Staphylococcus aureus; horizontal gene transfer; plasmid; superinfection exclusion; temperate bacteriophage; transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Mašlaňová I, Doškař J, Varga M, Kuntová L, Mužík J, Malúšková D, Růžičková V, Pantůček R. 2013. Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies. Environ Microbiol Rep 5:66–73. doi: 10.1111/j.1758-2229.2012.00378.x - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

