Epigenetic Changes Regulating Epithelial-Mesenchymal Plasticity in Human Trophoblast Differentiation
- PMID: 40643491
- PMCID: PMC12249213
- DOI: 10.3390/cells14130970
Epigenetic Changes Regulating Epithelial-Mesenchymal Plasticity in Human Trophoblast Differentiation
Abstract
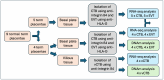
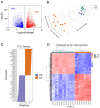
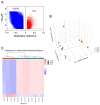
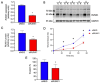
The phenotype of human placental extravillous trophoblast (EVT) at the end of pregnancy reflects both differentiation from villous cytotrophoblast (CTB) and later gestational changes, including loss of proliferative and invasive capacity. Invasion abnormalities are central to major obstetric pathologies, including placenta accreta spectrum, early onset preeclampsia, and fetal growth restriction. Characterization of the normal differentiation processes is, thus, essential for the analysis of these pathologies. Our gene expression analysis, employing purified human CTB and EVT cells, demonstrates a mechanism similar to the epithelial-mesenchymal transition (EMT), which underlies CTB-EVT differentiation. In parallel, DNA methylation profiling shows that CTB cells, already hypomethylated relative to non-trophoblast cell lineages, show further genome-wide hypomethylation in the transition to EVT. A small subgroup of genes undergoes gains of methylation (GOM), associated with differential gene expression (DE). Prominent in this GOM-DE group are genes involved in epithelial-mesenchymal plasticity (EMP). An exemplar is the transcription factor RUNX1, for which we demonstrate a functional role in regulating the migratory and invasive capacities of trophoblast cells. This analysis highlights epigenetically regulated genes acting to underpin the epithelial-mesenchymal plasticity characteristic of human trophoblast differentiation. Identification of these elements provides important information for the obstetric disorders in which these processes are dysregulated.
Keywords: DNA methylation; differentiation; epithelial–mesenchymal transition; trophoblast.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Update of
-
Epigenetic changes regulating the epithelial-mesenchymal transition in human trophoblast differentiation.bioRxiv [Preprint]. 2024 Jul 4:2024.07.02.601748. doi: 10.1101/2024.07.02.601748. bioRxiv. 2024. Update in: Cells. 2025 Jun 24;14(13):970. doi: 10.3390/cells14130970. PMID: 39005325 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

