Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically
- PMID: 40643521
- PMCID: PMC12248767
- DOI: 10.3390/cells14131000
Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically
Abstract
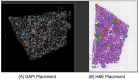
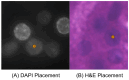
10X Xenium is an in situ spatial transcriptomics platform that enables single-cell and subcellular-level gene expression analysis. In Xenium data analysis, defining matched keypoints to align H&E and spatial transcriptomic images is critical for cross-referencing sequencing and histology. Currently, it is labor-intensive for domain experts to manually place keypoints to perform image registration in the Xenium Explorer software. We present Xenium-Align, a keypoint identification method that automatically generates keypoint files for image registration in Xenium Explorer. We validated our proposed method on 14 human kidney samples and one human skin Xenium sample representing healthy and diseased states, with expert manually marked results. These results show that Xenium-Align could generate accurate keypoints for automatically implementing image alignment in the Xenium Explorer software for spatial transcriptomics studies. Our future research aims to optimize the method's runtime efficiency and usability for image alignment applications.
Keywords: H&E image; Xenium technology; graph matching; image alignment; nucleus segmentation; spatial transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Spatial proteomics and transcriptomics characterization of tissue and multiple cancer types including decalcified marrow.Cancer Biomark. 2025 Jan;42(1):18758592241308757. doi: 10.1177/18758592241308757. Epub 2025 Mar 20. Cancer Biomark. 2025. PMID: 40109220
-
Leveraging a foundation model zoo for cell similarity search in oncological microscopy across devices.Front Oncol. 2025 Jun 18;15:1480384. doi: 10.3389/fonc.2025.1480384. eCollection 2025. Front Oncol. 2025. PMID: 40606969 Free PMC article.
-
Automated underwater plectropomus leopardus phenotype measurement through cylinder.Sci Rep. 2025 Jul 8;15(1):24461. doi: 10.1038/s41598-025-08863-w. Sci Rep. 2025. PMID: 40628812 Free PMC article.
-
MRI software and cognitive fusion biopsies in people with suspected prostate cancer: a systematic review, network meta-analysis and cost-effectiveness analysis.Health Technol Assess. 2024 Oct;28(61):1-310. doi: 10.3310/PLFG4210. Health Technol Assess. 2024. PMID: 39367754 Free PMC article.
-
Nivolumab for adults with Hodgkin's lymphoma (a rapid review using the software RobotReviewer).Cochrane Database Syst Rev. 2018 Jul 12;7(7):CD012556. doi: 10.1002/14651858.CD012556.pub2. Cochrane Database Syst Rev. 2018. PMID: 30001476 Free PMC article.
References
-
- Cook D.P., Jensen K.B., Wise K., Roach M.J., Dezem F.S., Ryan N.K., Zamojski M., Vlachos I.S., Knott S.R.V., Butler L.M., et al. A Comparative Analysis of Imaging-Based Spatial Transcriptomics Platforms. bioRxiv. 2023 doi: 10.1101/2023.12.13.571385. - DOI
-
- Janesick A., Shelansky R., Gottscho A.D., Wagner F., Williams S.R., Rouault M., Beliakoff G., Morrison C.A., Oliveira M.F., Sicherman J.T., et al. 10x Development Teams. High Resolution Mapping of the Tumor Microenvironment Using Integrated Single-Cell, Spatial and In Situ Analysis. Nat. Commun. 2023;14:8353. doi: 10.1038/s41467-023-43458-x. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

