Yoda1 Inhibits TGFβ-Induced Cardiac Fibroblast Activation via a BRD4-Dependent Pathway
- PMID: 40643546
- PMCID: PMC12249368
- DOI: 10.3390/cells14131028
Yoda1 Inhibits TGFβ-Induced Cardiac Fibroblast Activation via a BRD4-Dependent Pathway
Abstract
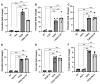
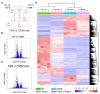
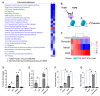
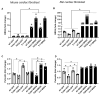
Fibrosis represents a pivotal pathological process in numerous diseases, characterized by excessive deposition of extracellular matrix (ECM) that disrupts normal tissue architecture and function. In the heart, cardiac fibrosis significantly impairs both structural integrity and functional capacity, contributing to the progression of heart failure. Central to this process are cardiac fibroblasts (CFs), which, upon activation, differentiate into contractile myofibroblasts, driving pathological ECM accumulation. Transforming growth factor-beta (TGFβ) is a well-established regulator of fibroblast activation; however, the precise molecular mechanisms, particularly the involvement of ion channels, remain poorly understood. Emerging evidence highlights the regulatory role of ion channels, including calcium-activated potassium (KCa) channels, in fibroblast activation. This study elucidates the role of ion channels and investigates the mechanism by which Yoda1, an agonist of the mechanosensitive ion channel Piezo1, modulates TGFβ-induced fibroblast activation. Using NIH/3T3 fibroblasts, we demonstrated that TGFβ-induced activation is regulated by tetraethylammonium (TEA)-sensitive potassium channels, but not by specific K⁺ channel subtypes such as BK, SK, or IK channels. Intriguingly, Yoda1 was found to inhibit TGFβ-induced fibroblast activation through a Piezo1-independent mechanism. Transcriptomic analysis revealed that Yoda1 modulates fibroblast activation by altering gene expression pathways associated with fibrotic processes. Bromodomain-containing protein 4 (BRD4) was identified as a critical mediator of Yoda1's effects, as pharmacological inhibition of BRD4 with JQ1 or ZL0454 suppressed TGFβ-induced expression of the fibroblast activation marker Periostin (Postn). Conversely, BRD4 overexpression attenuated the inhibitory effects of Yoda1 in both mouse and rat CFs. These results provide novel insights into the pharmacological modulation of TGFβ-induced cardiac fibroblast activation and highlight promising therapeutic targets for the treatment of fibrosis-related cardiac pathologies.
Keywords: BRD4; Periostin; Yoda1; cardiac fibroblast; fibroblast activation; ion channels; mechanosensitive channels.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Wang J., Hoshijima M., Lam J., Zhou Z., Jokiel A., Dalton N.D., Hultenby K., Ruiz-Lozano P., Ross J., Tryggvason K., et al. Cardiomyopathy associated with microcirculation dysfunction in laminin alpha4 chain-deficient mice. J. Biol. Chem. 2006;281:213–220. doi: 10.1074/jbc.M505061200. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

